WO2001014569A2 - Increasing the polysaccharide content in plants - Google Patents
Increasing the polysaccharide content in plants Download PDFInfo
- Publication number
- WO2001014569A2 WO2001014569A2 PCT/EP2000/007884 EP0007884W WO0114569A2 WO 2001014569 A2 WO2001014569 A2 WO 2001014569A2 EP 0007884 W EP0007884 W EP 0007884W WO 0114569 A2 WO0114569 A2 WO 0114569A2
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- plants
- plant
- dihydroorotase
- dna sequence
- coding
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Ceased
Links
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/63—Introduction of foreign genetic material using vectors; Vectors; Use of hosts therefor; Regulation of expression
- C12N15/79—Vectors or expression systems specially adapted for eukaryotic hosts
- C12N15/82—Vectors or expression systems specially adapted for eukaryotic hosts for plant cells, e.g. plant artificial chromosomes (PACs)
- C12N15/8241—Phenotypically and genetically modified plants via recombinant DNA technology
- C12N15/8242—Phenotypically and genetically modified plants via recombinant DNA technology with non-agronomic quality (output) traits, e.g. for industrial processing; Value added, non-agronomic traits
- C12N15/8243—Phenotypically and genetically modified plants via recombinant DNA technology with non-agronomic quality (output) traits, e.g. for industrial processing; Value added, non-agronomic traits involving biosynthetic or metabolic pathways, i.e. metabolic engineering, e.g. nicotine, caffeine
- C12N15/8245—Phenotypically and genetically modified plants via recombinant DNA technology with non-agronomic quality (output) traits, e.g. for industrial processing; Value added, non-agronomic traits involving biosynthetic or metabolic pathways, i.e. metabolic engineering, e.g. nicotine, caffeine involving modified carbohydrate or sugar alcohol metabolism, e.g. starch biosynthesis
- C12N15/8246—Non-starch polysaccharides, e.g. cellulose, fructans, levans
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/63—Introduction of foreign genetic material using vectors; Vectors; Use of hosts therefor; Regulation of expression
- C12N15/79—Vectors or expression systems specially adapted for eukaryotic hosts
- C12N15/82—Vectors or expression systems specially adapted for eukaryotic hosts for plant cells, e.g. plant artificial chromosomes (PACs)
- C12N15/8241—Phenotypically and genetically modified plants via recombinant DNA technology
- C12N15/8242—Phenotypically and genetically modified plants via recombinant DNA technology with non-agronomic quality (output) traits, e.g. for industrial processing; Value added, non-agronomic traits
- C12N15/8243—Phenotypically and genetically modified plants via recombinant DNA technology with non-agronomic quality (output) traits, e.g. for industrial processing; Value added, non-agronomic traits involving biosynthetic or metabolic pathways, i.e. metabolic engineering, e.g. nicotine, caffeine
- C12N15/8245—Phenotypically and genetically modified plants via recombinant DNA technology with non-agronomic quality (output) traits, e.g. for industrial processing; Value added, non-agronomic traits involving biosynthetic or metabolic pathways, i.e. metabolic engineering, e.g. nicotine, caffeine involving modified carbohydrate or sugar alcohol metabolism, e.g. starch biosynthesis
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/14—Hydrolases (3)
- C12N9/78—Hydrolases (3) acting on carbon to nitrogen bonds other than peptide bonds (3.5)
- C12N9/86—Hydrolases (3) acting on carbon to nitrogen bonds other than peptide bonds (3.5) acting on amide bonds in cyclic amides, e.g. penicillinase (3.5.2)
Definitions
- the present invention relates to the use of DNA sequences coding for a dihydroorotase for the production of plants with an increased polysaccharide or starch content, a process for the production of plants with increased polysaccharide or starch content by expressing a DNA sequence coding for a dihydroorotase , and the polysaccharide-overproducing plant itself.
- the invention further relates to a DNA sequence SEQ-ID No. 1 and with this hybridizing DNA sequence or homologous to the entire sequence or to partial sequences coding for a dihydroorotase from Solanum tuberosum.
- nucleotides are essential, particularly in rapidly growing tissues, and are therefore synthesized by multi-stage metabolic pathways. Pyrimidine nucleotides also play an important role as cofactors in vegetable carbohydrate metabolism. Up to 80% of the uridine nucleotides are present as UDP sugars, which are activated precursors for oligosaccharides or e.g. are required for cell wall synthesis (Wagner and Becker, 1992, Int. Rev. Cyt., 134, 1-84). UDP-glucose, for example, is the activated precursor for the synthesis of sucrose. Sucrose serves the plant as a transport form for glucose, the monomer of the starch, which is synthesized in the potato tubers for storage.
- the enzymes involved in starch biosynthesis are largely known.
- the sucrose made available from the leaves via the vascular system is mainly split into UDP-glucose and fructose by the enzyme sucrose synthase in a UDP-dependent reaction.
- the enzyme uridine diphosphoglucose pyrophosphorylase (UGPase) converts UDP-glucose to glucose-1-phosphate and UTP in a reaction dependent on pyrophosphate.
- ADP-glucose is used as an activated monomer for starch - synthesis by the enzyme starch synthase. This is provided by the enzyme ADP-glucose pyrophosphorylase (AGPase) from glucose-1-phosphate and ATP.
- AGPase ADP-glucose pyrophosphorylase
- the object of the invention was to increase the polysaccharide content in plant cells.
- the task was surprisingly achieved by expressing a gene coding for a dihydroorotase (DHO) in the plastids of transgenic plants.
- DHO dihydroorotase
- polysaccharides are preferably understood to mean starch, cellulose, hemicellulose, dextrans, pectins, mannans, galactans, xylans, inulins and fructans.
- polysaccharide preferably understood to mean starch, cellulose, hemicellulose, dextrans, pectins, mannans, galactans, xylans, inulins and fructans.
- other homogeneous or heterogeneous polysaccharides composed of glycosidically linked unmodified or modified monosaccharides of glucose and fructose are also understood to be the term polysaccharide.
- the transgenic polysaccharide overproducing plants are produced by transforming the plants with a construct containing a DHO gene.
- Tobacco, Arabidopsis thaliana, corn and potatoes were used as model plants for the production of polysaccharide overproducing plants.
- Genes coding for a dihydroorotase have previously been isolated from some organisms, inter alia from Saccharomyces cerevisiae (Genbank Acc. No .: X 07561), from Ustilago maydis (Genbank Acc. No.: X 63181), Arabidopis tha - liana (Genbank Acc. no .: AF 000146) and from E.coli (Genbank Acc. no .: X 04469).
- the invention relates to the use, for example, of a DNA sequence from E. coli (Genbank Acc. No. X04469), which codes for a DHO or its functional equivalents, for producing a plant with an increased content of polysaccharides.
- the nucleic acid sequence can be, for example, a DNA or cDNA sequence. Coding sequences suitable for insertion into an expression cassette are, for example, those which code for a DHO, are of homologous or heterologous origin and which preferably confer starch on the host the ability to overproduce polysaccharides.
- a DNA sequence suitable for insertion into an expression cassette is, for example, a DNA sequence SEQ-ID No. 1 and DNA sequence which hybridizes with it or which is homologous to the overall sequence or to partial sequences, for a dihydroorotase from solanum tuberosum.
- the expression cassettes also contain regulatory nucleic acid sequences which control the expression of the coding sequence in the host cell.
- an expression cassette comprises a polyadenylation signal upstream, ie at the 5 'end of the coding sequence, a promoter and downstream, ie at the 3' end, and optionally further regulatory elements which are associated with the coding sequence for the DHO gene are linked operatively.
- Operative linkage means the sequential Anord ⁇ of promoter, coding sequence, terminator and optionally other regulatory elements in such a way that each of the regulatory elements can fulfill its function in the expression of the coding sequence as intended.
- the preferred sequences for the operative linkage are targeting sequences to ensure subcellular localization in plastids.
- targeting sequences for ensuring subcellular localization in the mitochondrion, in the endoplasmic reticulum (ER), in the nucleus, in Olkorperchen or other compartments may if necessary, a ⁇ settable and translation enhancers such as the 5 '-Fuhrungssequenz from the tobacco mosaic virus ( Gallie et al., Nucl. Acids Res. 15 (1987), 8693-8711).
- the plant expression cassette can be incorporated into the Ta ⁇ bak transformation vector pBinAR-Hyg.
- Fig. 1 shows the tobacco transformation vectors pBinAR-Hyg with 35S promoter (A) or pBinAR-Hyg with seed-specific promoter Phaseolin 796 (B):
- HPT hygromycin phosphotransferase
- OCS octopine synthase terminator
- PNOS nopaline synthase promoter
- any promoter which can control the expression of foreign genes in plants is suitable as promoters of the expression cassette.
- a plant promoter or a plant virus-derived promoter is preferably used.
- the CaMV 35S promoter is particularly preferred
- this promoter contains different recognition sequences for transcriptional effectors, which in their entirety lead to permanent and constitutive expression of the introduced gene (Benfey et al., EMBO J. 8 (1989),
- the expression cassette can also contain a chemically inducible promoter, which controls the expression of the exogenous DHO gene in the plant at a specific point in time.
- Such promoters as e.g. the PRPl promoter (Ward et al., Plant. Mol. Biol. 22 (1993), 361-366), a promoter inducible by salicylic acid (WO 95/19443), one inducible by benzenesulfonamide (EP-A 388186 ), a tetracycline-inducible (Gatz et al., (1992) Plant J. 2, 397-404)
- promoters inducible by abscisic acid (EP-A 335528) or promoters inducible by ethanol or cyclohexanone (WO 93/21334) may include be used.
- promoters are particularly preferred which ensure expression in tissues or parts of plants in which, for example, the biosynthesis of starch or its precursors takes place. Promoters that ensure leaf-specific expression should be mentioned in particular.
- the promoter of the cytosolic FBPase from potatoes or the ST-LSI 35 promoter from potatoes should be mentioned (Stockhaus et al., EMBO J. 8 (1989), 2445-245).
- the expression cassette can therefore, for example, be a seed-specific promoter (preferably the phaseolin promoter (US 5504200), the USP- (Baumlein, H. et al., Mol. Gen.
- An expression cassette is produced by fusing a suitable promoter with a suitable DHO-DNA sequence and preferably a DNA inserted between the promoter and DHO-DNA sequence, which codes for a chloroplast-specific transit peptide, and a polyadenylation signal according to common recombination and cloning techniques as described, for example, in T. Maniatis, EF Fritsch and J. Sambrook, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989) and in T.J. Silhavy, M.L. Berman and L.W. Inquist, Experiments with Gene Fusions, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1984) and in Ausubel, F.M. et al. , Current Protocols in Molecular Biology, Greene Publishing Assoc. and Wiley-Interscience (1987).
- Transit peptides are preferred for the chloroplasts, which are cleaved enzymatically from the DHO part after translocation of the DHO gene into the chloroplasts.
- Particularly preferred is the transit peptide derived from the plastic DHO or a functional equivalent of this transit peptide (e.g. the transit peptide of the Rubisco small subunit or the ferredoxin NADP oxidoreductase).
- DNA sequences from three cassettes of the plastid transit peptide of potato plastid transketolase in three reading frames are particularly preferred as Kpnl / BamHI fragments with an ATG codon in the Ncol interface:
- the inserted nucleotide sequence coding for a DHO can be produced synthetically or obtained naturally or contain a mixture of synthetic and natural DNA components, as well as consist of different heterologous DHO gene sections of different organisms.
- synthetic nucleotide sequences with codons are generated which are preferred by plants. These codons preferred by plants can be determined from codons with the highest protein frequency, which are expressed in most interesting plant species.
- various DNA fragments can be manipulated in order to obtain a nucleotide sequence which expediently reads in the correct direction and which is equipped with a correct reading frame.
- adapters or linkers can be attached to the fragments.
- the promoter and terminator regions can expediently be provided in the transcription direction with a linker or polylinker which contains one or more restriction sites for the insertion of this sequence.
- the linker has 1 to 10, usually 1 to 8, preferably 2 to 6, restriction sites.
- the linker has a size of less than 100 bp within the regulatory areas, often less than 60 bp, but at least 5 bp.
- the promoter can be native or homologous as well as foreign or heterologous to the host plant.
- the expression cassette contains in the 5 '-3' transcription direction the promoter, a DNA sequence which codes for a DHO gene and a region for the transcriptional termination. Different termination areas are interchangeable.
- Preferred polyadenylation signals are plant polyadenylation signals, preferably those which essentially correspond to T-DNA polyadenylation signals from Agrobacterium tumefaciens, in particular gene 3 of T-DNA (octopine synthase) of the Ti plasmid pTiACH5 (Gielen et al., EMBO J. 3 (1984), 835 ff) or functional equivalents.
- An expression cassette can contain, for example, a constitutive promoter (preferably the CaMV 35 S promoter), the LeB4 signal peptide, the gene to be expressed and the ER retention signal.
- a constitutive promoter preferably the CaMV 35 S promoter
- the amino acid sequence KDEL lysine, aspartic acid, glutamic acid, leucine
- KDEL lysine, aspartic acid, glutamic acid, leucine
- the fused expression cassette which codes for a DHO gene is preferably cloned into a vector, for example pBin19, which is suitable for transforming Agrobacterium tumefaciens.
- Agrobacteria transformed with such a vector can then be used in a known manner to transform plants, in particular crop plants, such as, for example, tobacco plants, for example by bathing wounded leaves or leaf pieces in an agrobacterial solution and then cultivating them in suitable media.
- the transformation of plants by agrobacteria is known, inter alia, from FF White, Vectors for Gene Transfer in Higher Plants; in Transgenic Plants, Vol. 1, Engineering and Utilization, edited by SD Kung and R. Wu, Academic Press, 1993, pp. 15-38. From the transformed cells of the wounded leaves or leaf pieces, transgenic plants can be regenerated in a known manner which contain a gene integrated in the expression cassette for the expression of a DHO gene included.
- an expression cassette is inserted as an insert into a recombinant vector whose vector DNA contains additional functional regulatory signals, for example sequences for replication or integration.
- additional functional regulatory signals for example sequences for replication or integration.
- Suitable vectors are inter alia in "Methods in Plant Molecular Biology and Biotechnology" (CRC Press), Chap. 6/7, pp. 71-119 (1993).
- Cloning techniques allow the expression cassettes to be cloned into suitable vectors that allow them to multiply, for example in E. coli.
- suitable cloning vectors include pBR332, pUC series, M13mp series and pACYC184.
- Binary vectors which can replicate both in E. coli and in agrobacteria are particularly suitable.
- the invention further relates to the use of an expression cassette containing DNA sequences coding for a DHO gene or DNA sequences hybridizing therewith for the transformation of plants, cells, tissues or parts of plants.
- the aim of the use is to increase the content of polysaccharides, preferably starch, in plants.
- the expression of the DHO gene can take place specifically in the leaves, in the seeds, in the tubers or in other parts of the plant.
- Such polysaccharide-overproducing transgenic plants, their reproductive material, and their plant cells, tissue or parts are a further subject of the present invention.
- the expression cassette containing a DHO gene sequence according to the invention can also be used to transform bacteria, cyanobacteria, yeasts, filamentous fungi and algae with the aim of increasing the content of polysaccharides, preferably starch.
- transformation The transfer of foreign genes into the genome of a plant is called transformation.
- the methods described for the transformation and regeneration of plants are used
- Plant tissues or plant cells used for transient or stable transformation are used for transient or stable transformation. Suitable methods are the protoplast transformation by polyethylene glycol-induced DNA uptake, the biolistic method with the gene cannon - the so-called particle bombardment method, electroporation, the incubation of dry embryos in DNA-containing solution, microinjection and the gene transfer mediated by Agrobacterium. The methods mentioned are described, for example, in B. Jenes et al., Techniques for Gene Transfer, in: Transgenic Plants, Vol. 1, Engineering and Utilization, edited by SD Kung and R. Wu, Academic Press (1993), 128-143 and in Potrykus, Annu. Rev. Plant Physiol. Plant Molec. Biol. 42 (1991), 205-225).
- the construct to be expressed is preferably cloned into a vector which is suitable for transforming Agrobacterium tumefaciens, for example pBin19 (Bevan et al., Nucl. Acids Res. 12 (1984), 8711).
- Agrobacteria transformed with an expression cassette can also be used in a known manner to transform plants, in particular crop plants, such as cereals, maize, oats, rye, barley, wheat, soybeans, rice, cotton, sugar beet, canola, sunflower, flax, hemp, potatoes, Tobacco, tomato, rapeseed, tapioca, cassava, arrowroot, alfalfa, lettuce and the various tree, nut and wine species can be used, for example by bathing wounded leaves or leaf pieces in an agrobacterial solution and then cultivating them in suitable media.
- crop plants such as cereals, maize, oats, rye, barley, wheat, soybeans, rice, cotton, sugar beet, canola, sunflower, flax, hemp, potatoes, Tobacco, tomato, rapeseed, tapioca, cassava, arrowroot, alfalfa, lettuce and the various tree, nut and wine species can be used, for example by bathing wounded leaves or leaf pieces
- Functionally equivalent sequences which code for a DHO gene are those sequences which, despite a different nucleotide sequence, still have the desired functions. Functional equivalents thus include naturally occurring variants of the sequences described herein as well as artificial, e.g. Artificial nucleotide sequences obtained by chemical synthesis and adapted to the codon use of a plant.
- a functional equivalent is understood to mean, in particular, natural or artificial mutations of an originally isolated sequence coding for a DHO, which furthermore show the desired function. Mutations include substitutions, additions, deletions, exchanges or insertions of one or more nucleotide residues.
- the present invention also encompasses those nucleotide sequences which are obtained by modifying the DHO nucleotide sequence. The aim of such a modification can, for example, be to further narrow down the coding sequence contained therein or, for example, also to insert further restriction enzyme interfaces.
- Functional equivalents are also those variants whose function is weakened or enhanced compared to the original gene or gene fragment.
- artificial DNA sequences are suitable as long as, as described above, they impart the desired property, for example increasing the starch content in the plant by overexpressing the DHO gene in crop plants.
- Such artificial DNA sequences can be, for example, by Ruckuber
- Coding DNA sequences which are obtained by back-translating a polypeptide sequence according to the codon usage specific for the host plant are particularly suitable.
- the specific codon usage can be
- a person skilled in the art can easily determine 20 genetic methods by means of computer evaluations of other known genes of the plant to be transformed.
- Suitable equivalent nucleic acid sequences include 25 sequences which code for fusion proteins, part of the fusion protein being a DHO polypeptide or a functionally equivalent part thereof.
- the second part of the fusion protein can e.g. be another polypeptide with enzymatic activity or an antigenic polypeptide sequence that can be used to detect DHO expression (e.g. myc-tag or his-tag).
- this is preferably a regulatory protein sequence, such as e.g. a signal or transit peptide that directs the DHO protein to the desired site of action.
- increasing the polysaccharide content means, for example, the artificially acquired ability of an increased starch biosynthesis performance by functional overexpression of the DHO gene in the plant compared to the non-genetically modified plant for at least one period
- Starke's biosynthesis site for example, is generally leaf tissue, so that leaf-specific expression of the DHO gene makes sense.
- the starch bio-synthesis must not be restricted to the leaf tissue, but also in all other parts of the plant - for example in fatty seeds or in the tubers - tissue-specific.
- constitutive expression of the exogenous DHO gene is advantageous.
- inducible expression may also appear desirable.
- the effectiveness of the expression of the transgenically expressed DHO gene can be determined, for example, in vitro by propagation of the shoot meristem.
- a change in the type and level of expression of the DHO gene and its effect on the polysaccharide biosynthesis performance on test plants can be tested in greenhouse experiments.
- the invention also relates to transgenic plants transformed with an expression cassette containing a DHO gene sequence or DNA sequences hybridizing therewith, and transgenic cells, tissues, parts and propagation material of such plants.
- Transgenic crop plants such as e.g. Barley, wheat, rye, oats, corn, soybeans, rice, cotton, sugar beet, canola, sunflower, flax, hemp, potato, tobacco, tomato, rapeseed, tapioca, cassava, arrowroot, alfalfa, lettuce and the various tree nuts - and wine species.
- Plants in the sense of the invention are mono- and dicotyledonous plants or algae.
- the sequencing of recombinant DNA molecules was carried out with a laser fluorescence DNA sequencer from ABI according to the method of Sanger (Sanger et al. (1977) Proc. Natl. Acad. Sci. USA74, 5463-5467). Fragments resulting from a polymerase chain reaction were sequenced and checked in order to avoid polymerase errors in constructs to be expressed.
- RNA from plant tissues was, as in Logemann et al. (1987, Anal. Biochem. 163, 21). For the analysis, 20 ⁇ g RNA was separated in a 1.5% agarose gel containing formaldehyde and transferred to nylon membranes (Hybond, Amersham). The detection of specific transcripts was carried out as described in Aminos (1986, Anal. Biochem. 152, 304). The cDNA fragments used as a probe were radioactively labeled with a random primed DNA labeling kit (Boehringer, Mannheim) and hybridized according to standard methods (see Hybon user references, Amersham). Hyridization signals were visualized by autoradiography using X-OMAT AR films from Kodak.
- Potato plants (Solanum tuberosu L. cv.Desiree, Saatzucht Fritz Lange, Bad Schwartau) were grown in growth chambers (irradiance: 350 ⁇ mol photons ⁇ r ⁇ s -1 , 14 h / 10 h day / night rhythm, temperature: 20 ° C, 50 % relative humidity) in 3 1 pots on earth (with 100 g "Hakaphos green" [BASF-AG,
- Tuber disks with a thickness of 2 mm and a diameter of 8 mm were prepared as described in Geigenberger et al. (1997, Planta 201, 502-518). After washing three times with 10 mM 2- (N- morpholino) -ethane-sulfonic acid (Mes) (pH 6.5; KOH) the disks were incubated in 100 ml Erlenmeyer flasks at 90 rpm in the appropriate medium (8 disks in 4 ml).
- Mes 2- (N- morpholino) -ethane-sulfonic acid
- the tuber slices were extracted with 80% (v / v) ethanol (1 ml for 2 slices) and re-extracted in three subsequent steps (80% (v / v) ethanol, 50% (v / v) ethanol, H 2 0).
- the combined supernatants were dried in a stream of air at 47 ° C. and taken up in 1 ml of H0.
- This soluble fraction was, as in Quick et al. (1989, Planta 177, 536-546) separated into neutral, basic and acidic fractions by ion exchange chromatography. After freeze-drying, the neutral fraction was taken up in 100 ⁇ l of H 2 O and analyzed by means of thin layer chromatography (Geigenberger et al.
- Orotate and Uridm are precursors to uridine nucleotides.
- the question should be examined whether feeding with orotate or uridine has an influence on the nucleotide content in tuber disks.
- tuber slices of 10-week-old potato plants were incubated for 3 hours in the presence of 1 mM glucose and the corresponding uridine nucleotide precursors. The nucleotide contents were then measured.
- Figure 2 shows the nucleotide concentration in freshly prepared potato tuber slices from growing tubers of 10-week-old plants with or without feeding various nucleotide precursors (incubation for 3 hours in the presence of 10 mM Mes-KOH (pH 6.5), 300 mM mannitol) and 1 mM glucose Compared to non-incubated samples, the total uridine nucleotide content (UDPGlc + UTP + UDP; UMP was negligible) decreased by 30-40% after incubation with 1 mM glucose (Fig.
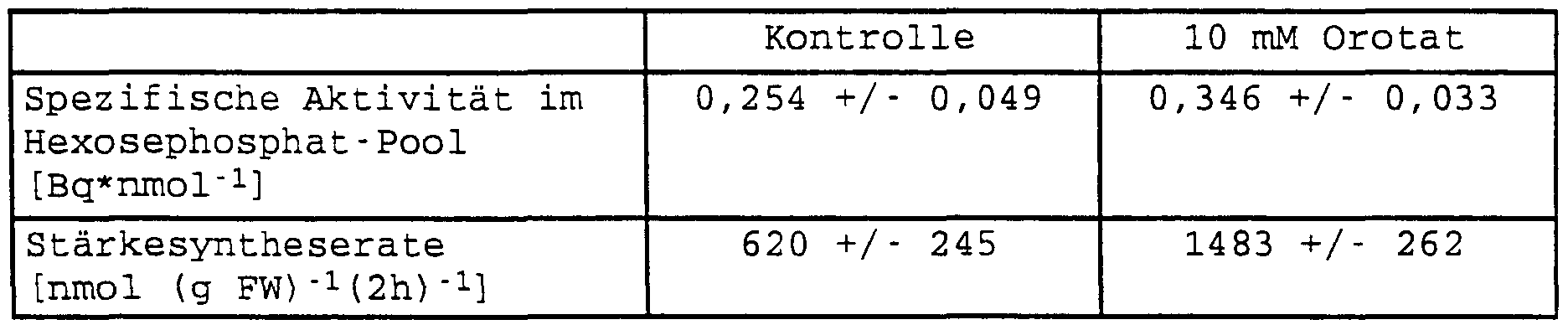
- tuber slices were incubated with 100 mM 14 C-sucrose in the presence and in the absence of 10 mM orotate. As in the presence of glucose, feeding with orotate led to an increase in uridine nucleotide concentrations without affecting adenylate and guanylate concentrations.
- FIG. 3 shows the nucleotide concentration in freshly prepared potato slices of growing tubers of 10-week-old plants without or with feeding 10 mM orotate (incubation for 3 hours in the presence of 10 mM Mes-KOH (pH 6.5) and 100 mM sucrose)
- Figure 4 shows the metabolism of 1 C - sucrose freshly prepared potato tuber slices from growing tubers of plants 10 weeks old without or with feeding of 10 mM orotate (incubation for 90 minutes in the presence of 100 mM sucrose. Subsequent addition of 1 C - sucrose ( 1.1 kBq ⁇ mol " 1 ) and incubation for a further 2 hours). Orotat led to a slight increase in the intake of 14 C-sucrose (Fig.
- Leaf disks of sterile plants were incubated in a Petri dish with a 1:50 agrobacterial dilution for 5-10 minutes. This was followed by a 2-day incubation in the dark at 25 ° C. on 2MS medium with 0.8% Bacto agar. e The cultivation was continued after 2 days with 16 hours of light / 8 hours of darkness and in a weekly rhythm on MS medium with 500 mg / 1 claforan (cefotaxime sodium), 50 mg / 1 Kanamycin, 1 mg / 1 benzylaminopurine (BAP), 0.2 mg / 1 naphthylacetic acid and 1.6 g / 1 glucose. Growing shoots were transferred to MS medium with 2% sucrose, 250 mg / 1 Claforan and 0.8% Bacto agar.
- the resulting 36 cDNA clones code for a polypeptide with homology to dihydroorotases from other organisms.
- the homology was obtained with the BLASTP program. (Altschul et al., Nucleic Acids Res. (1997) 25, 3389-3402). Accordingly, the protein is 78% identical to Arabidopsis thaliana dihydroorotase,
- the plasmid is called pBSSK-pyrCSt5.
- the cDNA (see SEQ ID No. 1) has an open reading frame of 1046 base pairs with a stop codon in position 1047-1049.
- the amino acid sequence begins with the third base in the reading frame and can be translated into a 348 amino acid polypeptide (see SEQ-ID No. 2). This corresponds to the length of prokaryotic dihydroorotase coding sequences.
- a clone coding for dihydroorotase was obtained from potato via the functional complementation of an E. coli mutant.
- the mutant CGSC5152 (CS101-2U5) of the E. coli Genetic Stock Center was used, which carries a mutation in the pyrC gene locus coding for a dihydroorotase.
- the complementation was carried out by electrotransformation of competent cells of the CGSC5152 strain with a cDNA bank in the vector plasmid pBS SK-.
- the underlying Lambda ZAPII bank (Stratagene) was cloned undirected using EcoRI / Notl linkers according to standard regulations.
- the RNA template for the cDNA was isolated from sink leaves of potato (small 1 cm leaflet from 10 week old potato plants harvested in a greenhouse).
- the transformed E. coli cells were plated on minimal medium M9 (Sambrook et al., 1989 see above), which additionally contained methionine (20 mg / 1), ampicillin (100 mg / 1) and IPTG (2.5 mM). A total of 4 micrograms of the bank were transformed in 8 approaches miert and 36 clones could be obtained, which follow
- a cDNA was produced which codes for an enzyme with dihydroorotase activity from potato which was fused to a signal sequence leading to the import of the protein into the plastids (taken from an enzyme with tranketolase activity from tobacco).
- the oligonucleotides 5 '-GTCGACATGGAGCTCTCAATCACACAACC-3' and... Were first of all determined using the pBSSK-pyrCSt5 cDNA
- PCR polymerase chain reaction
- Annealing temperature 50 ° C, 45 sec
- Denaturation temperature 95 ° C, 45 sec.
- Elongation temperature 72 ° C, 120 sec
- the fragment of approximately 1.1 kbp obtained was ligated into the vector pBluescript SK- (Stratagene) which had been cleaved with EcoRV.
- a clone K4 was identified by control cleavage, the insert of which can be excised in full length by Sall (1118 bp). The insert K4 was completely sequenced to rule out polymerase errors.
- a transfer vector was generated for the transformation of plants by ligating the 1118 bp Sall fragment from K4 into the vector pTK-TP-BinAR9 cleaved with Sall (R. Badur, 1998 doctoral thesis, University of Göttingen). The orientation of the insert was checked by cleavage with Kpnl (a fragment of approx. 980 bp resulted). In this way, the reading frame of the potato dihydroorotase was fused to a plastid transit peptide consisting of the N-terminal 60 amino acids achieved the tobacco transketolase (Genbank Acc. # CAA03393) (construct K5).
- the fused cDNA sequence is under the control of the cauliflower mosaic virus 35S promoter and the octopine synthase terminator from Agrobacterium tumefaciens.
- the construct K5 was used to transform tobacco, Arabidopsis thaliana and potato plants.
- Arabidopsis thaliana was transformed as in Bechtold, N., Ellis, J. and Pelletier, G. in Planta, Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants, C.R. Acad. Be. Paris, Life Sciences 316 (1993), 1194-1199.
- a cDNA was produced which codes for an enzyme with dihydroorotase activity from E. coli which was fused to a signal sequence leading to the import of the protein into the plastids (taken from an enzyme with tranketolase activity from tobacco).
- the oligonucleotides 5'-GTCGACAT-GACTGCACCATCCCAGG-3 'and 5' -CGATTTTTATTGTTTAACGGACC-3 'for a polymerase chain reaction (PCR) were first derived using the cDNA for the dihydroorotase from E. coli (Genbank Acc. No. X04469).
- a Sall- was identified by PCR with genomic DNA from E.coli XL-1 blue as a template.
- the reaction mixtures contained approx. 1 ng / ⁇ l of template DNA, 0.5 ⁇ M of the oligonucleotides and, 200 ⁇ M deoxy nucleotides (Pharmacia), 50 mM KCl, 10 mM Tris-HCl (pH 8.3 at 25 ° C. 1.5 mM MgCl) and 0.02 U / ⁇ l Pwo polymerase (Boehringer Mannheim) and were incubated in a PCR machine from Perkin Elmer with the following temperature program:
- Annealing temperature 50 ° C, 45 sec.
- Denaturation temperature 95 ° C, 45 sec
- Elongation temperature 72 ° C, 120 sec
- the 1059 bp fragment obtained was ligated into the vector pBluescript SK- (Stratagene), which had been split with EcoRV.
- a clone was identified by control cleavage, the insert of which can be excised in full length by Sall (1059 bp + 18 bp of the "multiple cloning site" of the vector).
- the 1077 bp Sall fragment from Kl was ligated into the expression vector pQE-9 (Quiagen). The correct orientation of the fragment was checked by restriction cleavage with BamHI.
- the pyrC E. coli mutant CGSC # 5152 (E. coli genetic stock center, York) was transformed with the construct K2 obtained. The transformants grew on M9 minimal media with 20 mg / l methionine without uridine, while mutants transformed with the empty pQE-9 vector showed no growth under these conditions.
- a transfer vector was generated for the transformation of plants by ligating the 1077 bp Sall fragment from Kl into the vector pTK-TP-BinAR9 cleaved with Sall (R. Badur, 1998 doctoral thesis, University of Göttingen). In this way a fusion of the reading frame of the dihydroorotase from E. coli to a plastid transit peptide, consisting of the N-terminal 60 amino acids of the transketolase from tobacco (Genbank Acc. # CAA03393) was achieved (construct K3, Fig. 5). The fused cDNA sequence is under the control of the cauliflower mosaic virus 35S promoter and the octopine synthase terminator from Agrobacterium tumefaciens.
- the construct K3 was used to transform tobacco, Arabidopsis thaliana and potato plants.
- Regenerated shoots were obtained on 2MS medium with kanamycin and claforan, transferred to soil after rooting and after cultivation for two weeks in a climatic chamber or in the greenhouse (as described above) for dihydroorotase expression Northern blot analysis examined. Lines with increased RNA levels of dihydroorotase were examined for altered metabolite and starch contents in leaf tissues or tubers. An increased uridine nucleotide content and an increased starch content were found in the transgenic lines compared to untransformed control plants.
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Genetics & Genomics (AREA)
- Engineering & Computer Science (AREA)
- Biotechnology (AREA)
- Chemical & Material Sciences (AREA)
- Molecular Biology (AREA)
- Zoology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Wood Science & Technology (AREA)
- Organic Chemistry (AREA)
- Biomedical Technology (AREA)
- General Engineering & Computer Science (AREA)
- Nutrition Science (AREA)
- Microbiology (AREA)
- Biochemistry (AREA)
- General Health & Medical Sciences (AREA)
- Biophysics (AREA)
- Cell Biology (AREA)
- Physics & Mathematics (AREA)
- Plant Pathology (AREA)
- Medicinal Chemistry (AREA)
- Enzymes And Modification Thereof (AREA)
- Breeding Of Plants And Reproduction By Means Of Culturing (AREA)
- Polysaccharides And Polysaccharide Derivatives (AREA)
Abstract
Description
Erhöhung des Polysaccharidgehaltes in PflanzenIncreasing the polysaccharide content in plants
Beschreibungdescription
Die vorliegende Erfindung betrifft die Verwendung von DNA-Sequenzen codierend für ein Dihydroorotase zur Herstellung von Pflanzen mit erhöhtem Polysaccharid- bzw. Stärkegehalt, ein Verfahren zur Herstellung von Pflanzen mit erhöhtem Polysaccharid bzw. Stärke- gehalt durch Expression einer DNA-Sequenz codierend für eine Dihydroorotase, sowie die derart hergestellte Polysaccharide-über- produzierende Pflanze selbst. Weiterhin betrifft die Erfindung eine DNA- Sequenz SEQ-ID No . 1 und mit dieser hybridisierende oder zur Gesamtsequenz oder zu Teilsequenzen homologen DNA- Sequenz ko- dierend für eine Dihydroorotase aus Solanum tuberosum.The present invention relates to the use of DNA sequences coding for a dihydroorotase for the production of plants with an increased polysaccharide or starch content, a process for the production of plants with increased polysaccharide or starch content by expressing a DNA sequence coding for a dihydroorotase , and the polysaccharide-overproducing plant itself. The invention further relates to a DNA sequence SEQ-ID No. 1 and with this hybridizing DNA sequence or homologous to the entire sequence or to partial sequences coding for a dihydroorotase from Solanum tuberosum.
Pflanzen synthetisieren ihre Zellkomponenten unter Nutzung der Sonnenenergie aus Kohlendioxid, Wasser und anorganischen Salzen. Nukleotide sind als elementare Bestandteile der Nukleinsäuren DNA und RNA insbesondere in schnell wachsenden Geweben essentiell und werden daher durch mehrstufige Stoffwechselwege synthetisiert. Pyrimidin-Nukleotide spielen darüber hinaus eine wichtige Rolle als Kofaktoren im pflanzlichen KohlenhydratstoffWechsel . Bis zu 80 % der Uridinnukleotide liegen als UDP-Zucker vor, die als ak- tivierte Vorstufen für Oligosaccharide oder z.B. für die Zell- wandsynthese benötigt werden (Wagner und Becker, 1992, Int. Rev. Cyt., 134, 1-84). UDP-Glucose stellt beispielsweise die aktivierte Vorstufe zur Synthese der Sucrose dar. Sucrose dient der Pflanze als Transportform für Glucose, dem Monomer der Stärke, die in den Kartoffelknollen zur Speicherung synthetisiert wird.Plants synthesize their cell components from carbon dioxide, water and inorganic salts using solar energy. As elementary components of the nucleic acids DNA and RNA, nucleotides are essential, particularly in rapidly growing tissues, and are therefore synthesized by multi-stage metabolic pathways. Pyrimidine nucleotides also play an important role as cofactors in vegetable carbohydrate metabolism. Up to 80% of the uridine nucleotides are present as UDP sugars, which are activated precursors for oligosaccharides or e.g. are required for cell wall synthesis (Wagner and Becker, 1992, Int. Rev. Cyt., 134, 1-84). UDP-glucose, for example, is the activated precursor for the synthesis of sucrose. Sucrose serves the plant as a transport form for glucose, the monomer of the starch, which is synthesized in the potato tubers for storage.
Die an der Stärkebiosynthese beteiligten Enzyme sind weitgehend bekannt. In der Kartoffelknolle wird die über das vaskuläre System aus den Blättern zur Verfügung gestellte Saccharose haupt- sächlich durch das Enzym Sucrose-Synthase in einer UDP-abhängigen Reaktion in UDP-Glucose und Fructose gespalten. Das Enzym Uridin- Diphosphoglucosepyrophosphorylase (UGPase) wandelt die UDP-Glucose in einer von Pyrophosphat abhängigen Reaktion zu Glu- cose-1-Phosphat und UTP um. Als aktiviertes Monomer zur Stärke - Synthese durch das Enzym Stärke-Synthase dient ADP-Glucose. Dieses wird durch das Enzym ADP-Glucose-Pyrophosphorylase (AGPase) aus Glucose-1-Phosphat und ATP bereitgestellt.The enzymes involved in starch biosynthesis are largely known. In the potato tuber, the sucrose made available from the leaves via the vascular system is mainly split into UDP-glucose and fructose by the enzyme sucrose synthase in a UDP-dependent reaction. The enzyme uridine diphosphoglucose pyrophosphorylase (UGPase) converts UDP-glucose to glucose-1-phosphate and UTP in a reaction dependent on pyrophosphate. ADP-glucose is used as an activated monomer for starch - synthesis by the enzyme starch synthase. This is provided by the enzyme ADP-glucose pyrophosphorylase (AGPase) from glucose-1-phosphate and ATP.
In den letzten Jahren wurde auf verschiedene Weise versucht, den Stärkegehalt in transgenen Kartoffelpflanzen zu erhöhen. Im Hinblick auf dieses Ziel ohne Erfolg waren Ansätze zur Überexpression von Invertase aus Hefe (Sonnewald et al . 1997, Nature Biotechnology 15: 794-797) sowie die kombinierte Expression von Glucokinase und Invertase in Kartoffelknollen (Trethewey et al . 1995, Plant J. 15: 109-118). Als erfolgreiche Ansätze zur Erhöhung der Stärkesynthese stellten sich die Überexpression einer AGPase (Stark et al. 1992, Science 258: 287-292), einer Pyro- phosphatase aus E.coli (Geigenberger et al. 1998, Planta 205: 428-434) oder eines ADP/ATP-Translokators dar (Tjaden et al . 1998, Plant Journal 16: 531-540). Diese Ergebnisse reflektieren die Verschiedenartigkeit der für die Stärkesynthese limitierenden Faktoren.Various attempts have been made in recent years to increase the starch content in transgenic potato plants. In view of this goal, attempts to overexpress invertase from yeast were unsuccessful (Sonnewald et al. 1997, Nature Biotechnology 15: 794-797) and the combined expression of glucokinase and invertase in potato tubers (Trethewey et al. 1995, Plant J. 15: 109-118). Overexpression of an AGPase (Stark et al. 1992, Science 258: 287-292), a pyrophosphatase from E. coli (Geigenberger et al. 1998, Planta 205: 428-434) turned out to be successful approaches to increasing starch synthesis. or an ADP / ATP translocator (Tjaden et al. 1998, Plant Journal 16: 531-540). These results reflect the diversity of the factors limiting the starch synthesis.
Wenig ist zur Zeit bekannt zur Rolle der Pyrimidin-Konzentration sowie der Uridinnukleotid-Umsätze für die Sucrosespaltung und die Stärkesynthese in Kartoffelknollen. Studien von Merlo et al . (1993, J. Plant Physiol. 142: 392-402) erbrachten korellative Hinweise für eine parallele Regulation des Uridinnukleotidstoffwech- sels mit dem Sucrose- und Stärkestoffwechsel zeigen jedoch keinen Weg auf gezielt die Stärkebiosynthese in Pflanzen zu steigern.Little is currently known about the role of pyrimidine concentration and uridine nucleotide sales for sucrose cleavage and starch synthesis in potato tubers. Studies by Merlo et al. (1993, J. Plant Physiol. 142: 392-402) provided corellative indications for a parallel regulation of the uridine nucleotide metabolism with the sucrose and starch metabolism, however, show no way to specifically increase the starch biosynthesis in plants.
Aufgabe der Erfindung war es, den Polysaccharidgehalt in Pflanzenzellen zu erhöhen.The object of the invention was to increase the polysaccharide content in plant cells.
Die Aufgabe konnte überraschenderweise gelöst werden durch Expression eines Gens kodierend für eine Dihydroorotase (DHO) in den Piastiden transgener Pflanzen.The task was surprisingly achieved by expressing a gene coding for a dihydroorotase (DHO) in the plastids of transgenic plants.
Erfindungsgemäß werden unter Polysacchariden vorzugsweise Stärke, Cellulose, Hemicellulose, Dextrane, Pektine, Mannane, Galactane, Xylane, Inuline und Fructane verstanden. Aber auch andere homo- gene oder heterogene Polysaccharide aufgebaut aus glykosidisch miteinander verknüpften nicht modifizierten oder modifizierten Monosacchariden der Glucose und der Fructose werden unter dem Begriff Polysaccharid verstanden.According to the invention, polysaccharides are preferably understood to mean starch, cellulose, hemicellulose, dextrans, pectins, mannans, galactans, xylans, inulins and fructans. However, other homogeneous or heterogeneous polysaccharides composed of glycosidically linked unmodified or modified monosaccharides of glucose and fructose are also understood to be the term polysaccharide.
Die Herstellung der transgenen Polysaccharide überproduzierenden Pflanzen erfolgt durch Transformation der Pflanzen mit einem ein DHO-Gen enthaltenden Konstrukt. Als Modellpflanze für die Produktion von Polysaccharide-überproduzierenden Pflanzen wurden Tabak, Arabidopsis thaliana, Mais und Kartoffel eingesetzt.The transgenic polysaccharide overproducing plants are produced by transforming the plants with a construct containing a DHO gene. Tobacco, Arabidopsis thaliana, corn and potatoes were used as model plants for the production of polysaccharide overproducing plants.
Gene, die für eine Dihydroorotase kodieren, wurden bereits zu einem früheren Zeitpunkt aus einigen Organismen isoliert, u.a. aus Saccharomyces cerevisiae ( Genbank Acc. Nr.: X 07561 ), aus Ustilago maydis ( Genbank Acc. Nr. : X 63181 ) , Arabidopis tha- liana ( Genbank Acc. Nr.: AF 000146) und aus E.coli (Genbank Acc. Nr . : X 04469) . Gegenstand der Erfindung ist die Verwendung beispielsweise einer DNA-Sequenz aus E. coli ( Genbank Acc. Nr. X04469 ), die für eine DHO oder deren funktionelle Äquivalente kodiert, zur Herstellung einer Pflanze mit erhöhtem Gehalt an Polysacchariden. Die Nukleinsäuresequenz kann dabei z.B. eine DNA- oder cDNA-Sequenz sein. Zur Insertion in eine Expressionskassette geeignete kodierende Sequenzen sind beispielsweise solche, die für eine DHO kodieren, homologen oder heterologen Ursprungs sind und die dem Wirt die Fähigkeit zur Überproduktion von Polysacchariden vorzugsweise Stärke verleihen.Genes coding for a dihydroorotase have previously been isolated from some organisms, inter alia from Saccharomyces cerevisiae (Genbank Acc. No .: X 07561), from Ustilago maydis (Genbank Acc. No.: X 63181), Arabidopis tha - liana (Genbank Acc. no .: AF 000146) and from E.coli (Genbank Acc. no .: X 04469). The invention relates to the use, for example, of a DNA sequence from E. coli (Genbank Acc. No. X04469), which codes for a DHO or its functional equivalents, for producing a plant with an increased content of polysaccharides. The nucleic acid sequence can be, for example, a DNA or cDNA sequence. Coding sequences suitable for insertion into an expression cassette are, for example, those which code for a DHO, are of homologous or heterologous origin and which preferably confer starch on the host the ability to overproduce polysaccharides.
Zur Insertion in eine Expressionskassette geeignete DNA- Sequenz ist beispielsweise eine DNA-Sequenz SEQ-ID No.l und mit dieser hybridisierende oder zur Gesamtsequenz oder zu Teilsequenzen homologen DNA- Sequenz kodierend f r eine Dihydroorotase aus Sola- num tuberosum.A DNA sequence suitable for insertion into an expression cassette is, for example, a DNA sequence SEQ-ID No. 1 and DNA sequence which hybridizes with it or which is homologous to the overall sequence or to partial sequences, for a dihydroorotase from solanum tuberosum.
Die Expressionskassetten beinhalten außerdem regulative Nuklein- sauresequenzen, welche die Expression der kodierenden Sequenz in der Wirtszelle steuern. Gemäß einer bevorzugten Ausfuhrungsform umfaßt eine Expressionskassette stromaufw rts, d.h. am 5' -Ende der kodierenden Sequenz, einen Promotor und stromabwärts, d.h. am 3' -Ende, ein Polyadenylierungssignal und gegebenenfalls weitere regulatorische Elemente, welche mit der dazwischenliegenden ko- dierenden Sequenz für das DHO-Gen operativ verknüpft sind. Unter einer operativen Verknüpfung versteht man die sequenzielle Anord¬ nung von Promotor, kodierender Sequenz, Terminator und ggf. weiterer regulativer Elemente derart, daß jedes der regulativen Elemente seine Funktion bei der Expression der kodierenden Sequenz bestimmungsgemäß erfüllen kann. Die zur operativen Verknüpfung bevorzugten Sequenzen sind Targeting-Sequenzen zur Gewährleistung der subzellularen Lokalisation in Plastiden. Aber auch Targeting- Sequenzen zur Gewährleistung der subzellulären Lokalisation im Mitochondrium, im Endoplasmatischen Retikulum (ER) , im Zellkern, in Olkorperchen oder anderen Kompartimenten sind bei Bedarf ein¬ setzbar sowie Translationsverstärker wie die 5' -Fuhrungssequenz aus dem Tabak-Mosaik-Virus (Gallie et al., Nucl. Acids Res. 15 (1987) , 8693 -8711) .The expression cassettes also contain regulatory nucleic acid sequences which control the expression of the coding sequence in the host cell. According to a preferred embodiment, an expression cassette comprises a polyadenylation signal upstream, ie at the 5 'end of the coding sequence, a promoter and downstream, ie at the 3' end, and optionally further regulatory elements which are associated with the coding sequence for the DHO gene are linked operatively. Operative linkage means the sequential Anord ¬ of promoter, coding sequence, terminator and optionally other regulatory elements in such a way that each of the regulatory elements can fulfill its function in the expression of the coding sequence as intended. The preferred sequences for the operative linkage are targeting sequences to ensure subcellular localization in plastids. However, targeting sequences for ensuring subcellular localization in the mitochondrion, in the endoplasmic reticulum (ER), in the nucleus, in Olkorperchen or other compartments may if necessary, a ¬ settable and translation enhancers such as the 5 '-Fuhrungssequenz from the tobacco mosaic virus ( Gallie et al., Nucl. Acids Res. 15 (1987), 8693-8711).
Beispielhaft kann die pflanzliche Expressionskassette in den Ta¬ bak-Transformationsvektor pBinAR-Hyg eingebaut werden. Abb. 1 zeigt die Tabaktransformationsvektoren pBinAR-Hyg mit 35S-Promo- tor (A) bzw. pBinAR-Hyg mit samenspezifischem Promotor Phaseolin 796 (B) :For example, the plant expression cassette can be incorporated into the Ta ¬ bak transformation vector pBinAR-Hyg. Fig. 1 shows the tobacco transformation vectors pBinAR-Hyg with 35S promoter (A) or pBinAR-Hyg with seed-specific promoter Phaseolin 796 (B):
HPT: Hygromycin-Phosphotransferase OCS: Octopin-Synthase-Terminator PNOS : Nopalin-Synthase-Promotor außerdem sind solche Restriktionsschnittstellen eingezeichnet, die nur einmal den Vektor schneiden.HPT: hygromycin phosphotransferase OCS: octopine synthase terminator PNOS: nopaline synthase promoter, in addition, such restriction sites are shown that cut the vector only once.
5 Als Promotoren der Expressionskassette ist grundsätzlich jeder Promotor geeignet, der die Expression von Fremdgenen in Pflanzen steuern kann. Vorzugsweise verwendet man insbesondere einen pflanzlichen Promotor oder einen Promotor, der einem Pflanzenvirus entstammt. Insbesondere bevorzugt ist der CaMV 35S-Promotor5 In principle, any promoter which can control the expression of foreign genes in plants is suitable as promoters of the expression cassette. In particular, a plant promoter or a plant virus-derived promoter is preferably used. The CaMV 35S promoter is particularly preferred
10 aus dem Blumenkohl-Mosaik-Virus (Franck et al . , Cell 21 (1980), 285 - 294) . Dieser Promotor enthält bekanntlich unterschiedliche ErkennungsSequenzen für transkriptionale Effektoren, die in ihrer Gesamtheit zu einer permanenten und konstitutiven Expression des eingeführten Gens führen (Benfey et al . , EMBO J. 8 (1989),10 from the cauliflower mosaic virus (Franck et al., Cell 21 (1980), 285-294). As is known, this promoter contains different recognition sequences for transcriptional effectors, which in their entirety lead to permanent and constitutive expression of the introduced gene (Benfey et al., EMBO J. 8 (1989),
15 2195-2202) .15 2195-2202).
Die Expressionskassette kann auch einen chemisch induzierbaren Promotor enthalten, durch den die Expression des exogenen DHO- Gens in der Pflanze zu einem bestimmten Zeitpunkt gesteuert wer-The expression cassette can also contain a chemically inducible promoter, which controls the expression of the exogenous DHO gene in the plant at a specific point in time.
20 den kann. Derartige Promotoren wie z.B. der PRPl-Promotor (Ward et al., Plant. Mol. Biol. 22 (1993), 361-366), ein durch Salizylsäure induzierbarer Promotor (WO 95/19443), ein durch Benzenesul- fonamid-induzierbarer (EP-A 388186) , ein durch Tetrazyklin- induzierbarer (Gatz et al., (1992) Plant J. 2, 397-404), ein20 den can. Such promoters as e.g. the PRPl promoter (Ward et al., Plant. Mol. Biol. 22 (1993), 361-366), a promoter inducible by salicylic acid (WO 95/19443), one inducible by benzenesulfonamide (EP-A 388186 ), a tetracycline-inducible (Gatz et al., (1992) Plant J. 2, 397-404)
25 durch Abscisinsäure-induzierbarer (EP-A 335528) bzw. ein durch Ethanol- oder Cyclohexanon-induzierbarer (WO 93/21334) Promotor können u.a. verwendet werden.25 promoters inducible by abscisic acid (EP-A 335528) or promoters inducible by ethanol or cyclohexanone (WO 93/21334) may include be used.
Weiterhin sind insbesonders solche Promotoren bevorzugt, die die 30 Expression in Geweben oder Pflanzenteilen sicherstellen, in denen beispielsweise die Biosynthese von Stärke bzw. deren Vorstufen stattfindet. Insbesondere zu nennen sind Promotoren, die eine blattspezifische Expression gewährleisten. Zu nennen sind der Promotor der cytosolischen FBPase aus Kartoffel oder der ST-LSI 35 Promotor aus Kartoffel (Stockhaus et al., EMBO J. 8 (1989), 2445 - 245) .Furthermore, promoters are particularly preferred which ensure expression in tissues or parts of plants in which, for example, the biosynthesis of starch or its precursors takes place. Promoters that ensure leaf-specific expression should be mentioned in particular. The promoter of the cytosolic FBPase from potatoes or the ST-LSI 35 promoter from potatoes should be mentioned (Stockhaus et al., EMBO J. 8 (1989), 2445-245).
Mit Hilfe eines samenspezifischen Promotors konnte ein Fremdprotein stabil bis zu einem Anteil von 0,67 % des gesamten lösli-With the help of a seed-specific promoter, a foreign protein was stable up to a share of 0.67% of the total soluble
40 chen Samenproteins in den Samen transgener Tabakpflanzen expri - miert werden (Fiedler und Conrad, Bio/Technology 10 (1995) , 1090-1094) . Die Expressionskassette kann daher beispielsweise einen samenspezifischen Promotor (bevorzugt den Phaseolin- Promotor (US 5504200), den USP- (Baumlein, H. et al . , Mol. Gen.40 seed protein can be expressed in the seeds of transgenic tobacco plants (Fiedler and Conrad, Bio / Technology 10 (1995), 1090-1094). The expression cassette can therefore, for example, be a seed-specific promoter (preferably the phaseolin promoter (US 5504200), the USP- (Baumlein, H. et al., Mol. Gen.
45 Genet. (1991) 225 (3), 459 - 467) oder LEB4-Promotor (Fiedler und Conrad, 1995)), das LEB4-Signalpeptid, das zu exprimierende Gen und ein ER-Retentionssignal enthalten.45 Genet. (1991) 225 (3), 459-467) or LEB4 promoter (Fiedler and Conrad, 1995)), the LEB4 signal peptide, the gene to be expressed and an ER retention signal.
Die Herstellung einer Expressionskassette erfolgt durch Fusion eines geeigneten Promotors mit einer geeigneten DHO-DNA-Sequenz und vorzugsweise einer zwischen Promotor und DHO-DNA-Sequenz inserierten DNA, die für ein chloroplastenspezifisches Transitpep- tid kodiert, sowie einem Polyadenylierungssignal nach gängigen Rekombinations- und Klonierungstechniken, wie sie beispielsweise in T. Maniatis, E.F. Fritsch und J. Sambrook, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989) sowie in T.J. Silhavy, M.L. Berman und L.W. En- quist, Experiments with Gene Fusions, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1984) und in Ausubel, F.M. et al . , Current Protocols in Molecular Biology, Greene Publishing Assoc. and Wiley-Interscience (1987) beschrieben sind.An expression cassette is produced by fusing a suitable promoter with a suitable DHO-DNA sequence and preferably a DNA inserted between the promoter and DHO-DNA sequence, which codes for a chloroplast-specific transit peptide, and a polyadenylation signal according to common recombination and cloning techniques as described, for example, in T. Maniatis, EF Fritsch and J. Sambrook, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989) and in T.J. Silhavy, M.L. Berman and L.W. Inquist, Experiments with Gene Fusions, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1984) and in Ausubel, F.M. et al. , Current Protocols in Molecular Biology, Greene Publishing Assoc. and Wiley-Interscience (1987).
Insbesondere bevorzugt sind Sequenzen, die ein Targeting in Pla- stiden gewährleisten. Unter bestimmten Umständen kann auch ein targeting in die Vakuole, in das Mitochondrium, in das Endoplas- matische Retikulum (ER) oder durch ein Fehlen entsprechender operativer Sequenzen ein Verbleib im Kompartiment des Entstehens, dem Zytosol, wünschenswert sein (Kermode, Crit. Rev. Plant Sei. 15, 4 (1996) , 285-423) .Sequences which ensure targeting in plastids are particularly preferred. Under certain circumstances, targeting into the vacuole, into the mitochondrium, into the endoplasmic reticulum (ER) or due to the lack of corresponding operative sequences, it may be desirable to remain in the compartment of formation, the cytosol (Kermode, Crit. Rev. Plant Sci., 15: 4 (1996), 285-423).
Es können auch Expressionskassetten verwendet werden, deren DNA- Sequenz für ein DHO-Fusionsprotein kodiert, wobei ein Teil des Fusionsproteins ein Transitpeptid ist, das die Translokation des Polypeptides steuert. Bevorzugt sind für die Chloroplasten spezi- fische Transitpeptide, welche nach Translokation des DHO-Gens in die Chloroplasten vom DHO-Teil enzymatisch abgespalten werden. Insbesondere bevorzugt ist das Transitpeptid, das von der plasti- dären DHO oder einem funktioneilen Äquivalent dieses Transitpep- tids (z.B. dem Transitpeptid der kleinen Untereinheit der Rubisco oder der Ferredoxin NADP Oxidoreduktase) abgeleitet ist.Expression cassettes whose DNA sequence codes for a DHO fusion protein can also be used, part of the fusion protein being a transit peptide which controls the translocation of the polypeptide. Preferred transit peptides are preferred for the chloroplasts, which are cleaved enzymatically from the DHO part after translocation of the DHO gene into the chloroplasts. Particularly preferred is the transit peptide derived from the plastic DHO or a functional equivalent of this transit peptide (e.g. the transit peptide of the Rubisco small subunit or the ferredoxin NADP oxidoreductase).
Besonders bevorzugt sind DNA-Sequenzen von drei Kassetten des Plastiden-Transitpeptids der plastidären Transketolase aus Kartoffel in drei Leserastern als Kpnl/BamHI Fragmente mit einem ATG-Codon in der Ncol Schnittstelle:DNA sequences from three cassettes of the plastid transit peptide of potato plastid transketolase in three reading frames are particularly preferred as Kpnl / BamHI fragments with an ATG codon in the Ncol interface:
pTP09pTP09
KpnI_GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTCKpnI_GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC
CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGGGA TCC_BamHICCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGGGA TCC_BamHI
pTPlO KpnI_GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGCTG GATCC_BamHIpTPlO KpnI_GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGCTG GATCC_BamHI
pTPllpTPll
KpnI_GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGGGG ATCC_BamHIKpnI_GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGGGG ATCC_BamHI
Die inserierte Nukleotid-Sequenz kodierend für eine DHO kann synthetisch hergestellt oder natürlich gewonnen sein oder eine Mischung aus synthetischen und natürlichen DNA-Bestandteilen enthalten, sowie aus verschiedenen heterologen DHO-Genabschnitten verschiedener Organismen bestehen. Im allgemeinen werden synthetische Nukleotid-Sequenzen mit Kodons erzeugt, die von Pflanzen bevorzugt werden. Diese von Pflanzen bevorzugten Kodons können aus Kodons mit der höchsten Proteinhäufigkeit bestimmt werden, die in den meisten interessanten Pflanzenspezies exprimiert werden. Bei der Präparation einer Expressionskassette können verschiedene DNA-Fragmente manipuliert werden, um eine Nukleotid-Sequenz zu erhalten, die zweckmäßigerweise in der korrekten Rich- tung liest und die mit einem korrekten Leseraster ausgestattet ist. Für die Verbindung der DNA-Fragmente miteinander können an die Fragmente Adaptoren oder Linker angesetzt werden.The inserted nucleotide sequence coding for a DHO can be produced synthetically or obtained naturally or contain a mixture of synthetic and natural DNA components, as well as consist of different heterologous DHO gene sections of different organisms. In general, synthetic nucleotide sequences with codons are generated which are preferred by plants. These codons preferred by plants can be determined from codons with the highest protein frequency, which are expressed in most interesting plant species. When preparing an expression cassette, various DNA fragments can be manipulated in order to obtain a nucleotide sequence which expediently reads in the correct direction and which is equipped with a correct reading frame. To connect the DNA fragments to one another, adapters or linkers can be attached to the fragments.
Zweckmäßigerweise können die Promotor- und die Terminator-Regio- nen in Transkriptionsrichtung mit einem Linker oder Polylinker, der eine oder mehrere Restriktionsstellen für die Insertion dieser Sequenz enthält, versehen werden. In der Regel hat der Linker 1 bis 10, meistens 1 bis 8, vorzugsweise 2 bis 6 Restriktions- stellen. Im allgemeinen hat der Linker innerhalb der regulatori- sehen Bereiche eine Größe von weniger als 100 bp, häufig weniger als 60 bp, mindestens jedoch 5 bp. Der Promotor kann sowohl nativ bzw. homolog als auch fremdartig bzw. heterolog zur Wirtspflanze sein. Die Expressionskassette beinhaltet in der 5' -3 ' -Transkrip- tionsrich ung den Promotor, eine DNA-Sequenz die für ein DHO-Gen codiert und eine Region für die transkriptionale Termination. Verschiedene Terminationsbereiche sind gegeneinander beliebig austauschbar.The promoter and terminator regions can expediently be provided in the transcription direction with a linker or polylinker which contains one or more restriction sites for the insertion of this sequence. As a rule, the linker has 1 to 10, usually 1 to 8, preferably 2 to 6, restriction sites. In general, the linker has a size of less than 100 bp within the regulatory areas, often less than 60 bp, but at least 5 bp. The promoter can be native or homologous as well as foreign or heterologous to the host plant. The expression cassette contains in the 5 '-3' transcription direction the promoter, a DNA sequence which codes for a DHO gene and a region for the transcriptional termination. Different termination areas are interchangeable.
Ferner können Manipulationen, die passende Restriktionsschnitt - stellen bereitstellen oder die überflüssige DNA oder Restriktionsschnittstellen entfernen, eingesetzt werden. Wo Insertionen, Deletionen oder Substitutionen wie z.B. Transitionen und Trans - Versionen in Frage kommen, können in vitro-Mutagenese, "primerre- pair" , Restriktion oder Ligation verwendet werden. Bei geeigneten Manipulationen, wie z.B. Restriktion, "chewing-back" oder Auffüllen von Überhängen für "bluntends" , können komplementäre Enden der Fragmente für die Ligation zur Verfügung gestellt werden.Manipulations which provide suitable restriction sites or which remove unnecessary DNA or restriction sites can also be used. Where insertions, deletions or substitutions such as Transitions and trans versions can be used in vitro mutagenesis, "primer pair", restriction or ligation. With suitable manipulations, e.g. Restriction, "chewing-back" or filling of overhangs for "bluntends", complementary ends of the fragments can be provided for the ligation.
Von Bedeutung für den erfindungsgemäßen Erfolg kann u.a. das An- hängen des spezifischen ER-Retentionssignals SEKDEL sein (Schou- ten, A. et al . , Plant Mol. Biol. 30 (1996), 781 - 792), die durchschnittliche Expressionshöhe wird damit verdreifacht bis vervierfacht. Es können auch andere Retentionssignale, die natürlicherweise bei im ER lokalisierten pflanzlichen und tierischen Proteinen vorkommen, für den Aufbau der Kassette eingesetzt werden.Of importance for the success according to the invention can i.a. the attachment of the specific ER retention signal SEKDEL (Schouten, A. et al., Plant Mol. Biol. 30 (1996), 781-792), the average expression level is tripled to quadrupled. Other retention signals, which occur naturally in plant and animal proteins located in the ER, can also be used to construct the cassette.
Bevorzugte Polyadenylierungssignale sind pflanzliche Polyadeny- lierungssignale, vorzugsweise solche, die im wesentlichen T-DNA- Polyadenylierungssignale aus Agrobacterium tumefaciens, insbesondere des Gens 3 der T-DNA (Octopin Synthase) des Ti-Plasmids pTiACH5 entsprechen (Gielen et al . , EMBO J. 3 (1984), 835 ff) oder funktioneile Äquivalente.Preferred polyadenylation signals are plant polyadenylation signals, preferably those which essentially correspond to T-DNA polyadenylation signals from Agrobacterium tumefaciens, in particular gene 3 of T-DNA (octopine synthase) of the Ti plasmid pTiACH5 (Gielen et al., EMBO J. 3 (1984), 835 ff) or functional equivalents.
Eine Expressionskassette kann beispielsweise einen konstitutiven Promotor (bevorzugt den CaMV 35 S-Promotor) , das LeB4-Signalpep- tid, das zu exprimierende Gen und das ER-Retentionssignal enthalten. Als ER-Retentionssignal wird bevorzugt die Aminosäuresequenz KDEL (Lysin, Asparaginsäure, Glutaminsäure, Leucin) verwendet.An expression cassette can contain, for example, a constitutive promoter (preferably the CaMV 35 S promoter), the LeB4 signal peptide, the gene to be expressed and the ER retention signal. The amino acid sequence KDEL (lysine, aspartic acid, glutamic acid, leucine) is preferably used as the ER retention signal.
Vorzugsweise wird die fusionierte Expressionskassette, die für ein DHO-Gen kodiert, in einen Vektor, beispielsweise pBinl9, klo- niert, der geeignet ist, Agrobacterium tumefaciens zu transformieren. Mit einem solchen Vektor transformierte Agrobakterien können dann in bekannter Weise zur Transformation von Pflanzen, insbesondere von Kulturpflanzen, wie z.B. von Tabakpflanzen, verwendet werden, indem beispielsweise verwundete Blätter oder Blattstücke in einer Agrobakterienlösung gebadet und anschließend in geeigneten Medien kultiviert werden. Die Transformation von Pflanzen durch Agrobakterien ist unter anderem bekannt aus F.F. White, Vectors for Gene Transfer in Higher Plants; in Transgenic Plants, Vol. 1, Engineering and Utilization, herausgegeben von S.D. Kung und R. Wu, Academic Press, 1993, S. 15 - 38. Aus den transformierten Zellen der verwundeten Blätter bzw. Blattstücke können in bekannter Weise transgene Pflanzen regeneriert werden, die ein in die Expressionskassette integriertes Gen für die Ex- pression eines DHO-Gens enthalten.The fused expression cassette which codes for a DHO gene is preferably cloned into a vector, for example pBin19, which is suitable for transforming Agrobacterium tumefaciens. Agrobacteria transformed with such a vector can then be used in a known manner to transform plants, in particular crop plants, such as, for example, tobacco plants, for example by bathing wounded leaves or leaf pieces in an agrobacterial solution and then cultivating them in suitable media. The transformation of plants by agrobacteria is known, inter alia, from FF White, Vectors for Gene Transfer in Higher Plants; in Transgenic Plants, Vol. 1, Engineering and Utilization, edited by SD Kung and R. Wu, Academic Press, 1993, pp. 15-38. From the transformed cells of the wounded leaves or leaf pieces, transgenic plants can be regenerated in a known manner which contain a gene integrated in the expression cassette for the expression of a DHO gene included.
Zur Transformation einer Wirtspflanze mit einer für eine DHO kodierende DNA wird eine Expressionskassette als Insertion in einen rekombinanten Vektor eingebaut, dessen Vektor-DNA zusätzliche funktioneile Regulationssignale, beispielsweise Sequenzen für Re- plikation oder Integration enthält. Geeignete Vektoren sind unter anderem in "Methods in Plant Molecular Biology and Biotechnology" (CRC Press), Kap. 6/7, S. 71 - 119 (1993) beschrieben.To transform a host plant with a DNA coding for a DHO, an expression cassette is inserted as an insert into a recombinant vector whose vector DNA contains additional functional regulatory signals, for example sequences for replication or integration. Suitable vectors are inter alia in "Methods in Plant Molecular Biology and Biotechnology" (CRC Press), Chap. 6/7, pp. 71-119 (1993).
Unter Verwendung der oben zitierten Rekombinations- undUsing the recombination and
Klonierungstechniken können die Expressionskassetten in geeignete Vektoren kloniert werden, die ihre Vermehrung, beispielsweise in E. coli, ermöglichen. Geeignete Klonierungsvektoren sind u.a. pBR332, pUC-Serien, M13mp-Serien und pACYC184. Besonders geeignet sind binäre Vektoren, die sowohl in E. coli als auch in Agrobakterien replizieren können.Cloning techniques allow the expression cassettes to be cloned into suitable vectors that allow them to multiply, for example in E. coli. Suitable cloning vectors include pBR332, pUC series, M13mp series and pACYC184. Binary vectors which can replicate both in E. coli and in agrobacteria are particularly suitable.
Ein weiterer Gegenstand der Erfindung betrifft die Verwendung einer Expressionskassette enthaltend DNA-Sequenzen codierend für ein DHO-Gen oder mit diesen hybridisierende DNA-Sequenzen zur Transformation von Pflanzen, -zellen, -geweben oder Pflanzenteilen. Ziel der Verwendung ist die Erhöhung des Gehaltes an Polysacchariden vorzugsweise an Stärke in Pflanzen.The invention further relates to the use of an expression cassette containing DNA sequences coding for a DHO gene or DNA sequences hybridizing therewith for the transformation of plants, cells, tissues or parts of plants. The aim of the use is to increase the content of polysaccharides, preferably starch, in plants.
Dabei kann je nach Wahl des Promotors die Expression des DHO-Gens spezifisch in den Blättern, in den Samen, den Knollen oder anderen Teilen der Pflanze erfolgen. Solche Polysaccharide-überprodu- zierenden transgenen Pflanzen, deren Vermehrungsgut, sowie deren Pflanzenzellen, -gewebe oder -teile sind ei -weiterer Gegenstand der vorliegenden Erfindung.Depending on the choice of the promoter, the expression of the DHO gene can take place specifically in the leaves, in the seeds, in the tubers or in other parts of the plant. Such polysaccharide-overproducing transgenic plants, their reproductive material, and their plant cells, tissue or parts are a further subject of the present invention.
Die Expressionskassette enthaltend eine erfindungsgemäße DHO-Gen- sequenz kann darüberhinaus auch zur Transformation von Bakterien, Cyanobakterien, Hefen, filamentösen Pilzen und Algen mit dem Ziel einer Erhöhung des Gehaltes an Polysacchariden vorzugsweise an Stärke eingesetzt werden.The expression cassette containing a DHO gene sequence according to the invention can also be used to transform bacteria, cyanobacteria, yeasts, filamentous fungi and algae with the aim of increasing the content of polysaccharides, preferably starch.
Die Übertragung von Fremdgenen in das Genom einer Pflanze wird als Transformation bezeichnet. Es werden dabei die beschriebenen Methoden zur Transformation und Regeneration von Pflanzen ausThe transfer of foreign genes into the genome of a plant is called transformation. The methods described for the transformation and regeneration of plants are used
Pflanzengeweben oder Pflanzenzellen zur transienten oder stabilen Transformation genutzt. Geeignete Methoden sind die Protoplasten- transformation durch Polyethylenglykol-induzierte DNA-Aufnahme, das biolistische Verfahren mit der Genkanone - die sogenannte particle bombardment Methode, die Elektroporation, die Inkubation trockener Embryonen in DNA-haltiger Lösung, die Mikroinj ektion und der durch Agrobacterium vermittelte Gentransfer. Die genannten Verfahren sind beispielsweise in B. Jenes et al., Techniques for Gene Transfer, in: Transgenic Plants, Vol. 1, Engineering and Utilization, herausgegeben von S.D. Kung und R. Wu, Academic Press (1993), 128 - 143 sowie in Potrykus, Annu. Rev. Plant Phy- siol. Plant Molec. Biol. 42 (1991), 205 - 225) beschrieben.Plant tissues or plant cells used for transient or stable transformation. Suitable methods are the protoplast transformation by polyethylene glycol-induced DNA uptake, the biolistic method with the gene cannon - the so-called particle bombardment method, electroporation, the incubation of dry embryos in DNA-containing solution, microinjection and the gene transfer mediated by Agrobacterium. The methods mentioned are described, for example, in B. Jenes et al., Techniques for Gene Transfer, in: Transgenic Plants, Vol. 1, Engineering and Utilization, edited by SD Kung and R. Wu, Academic Press (1993), 128-143 and in Potrykus, Annu. Rev. Plant Physiol. Plant Molec. Biol. 42 (1991), 205-225).
Vorzugsweise wird das zu exprimierende Konstrukt in einen Vektor kloniert, der geeignet ist, Agrobacterium tumefaciens zu transformieren, beispielsweise pBinl9 (Bevan et al., Nucl. Acids Res . 12 (1984) , 8711) .The construct to be expressed is preferably cloned into a vector which is suitable for transforming Agrobacterium tumefaciens, for example pBin19 (Bevan et al., Nucl. Acids Res. 12 (1984), 8711).
Mit einer Expressionskassette transformierte Agrobakterien können ebenfalls in bekannter Weise zur Transformation von Pflanzen, insbesondere von Kulturpflanzen, wie Getreide, Mais, Hafer, Roggen, Gerste, Weizen, Soja, Reis, Baumwolle, Zuckerrübe, Canola, Sonnenblume, Flachs, Hanf, Kartoffel, Tabak, Tomate, Raps, Ta- pioka, Maniok, Pfeilwurz, Alfalfa, Salat und den verschiedenen Baum-, Nuß- und Weinspezies, verwendet werden, z.B. indem verwundete Blätter oder Blattstücke in einer Agrobakterienlösung gebadet und anschließend in geeigneten Medien kultiviert werden.Agrobacteria transformed with an expression cassette can also be used in a known manner to transform plants, in particular crop plants, such as cereals, maize, oats, rye, barley, wheat, soybeans, rice, cotton, sugar beet, canola, sunflower, flax, hemp, potatoes, Tobacco, tomato, rapeseed, tapioca, cassava, arrowroot, alfalfa, lettuce and the various tree, nut and wine species can be used, for example by bathing wounded leaves or leaf pieces in an agrobacterial solution and then cultivating them in suitable media.
Funktioneil äquivalente Sequenzen, die für ein DHO-Gen kodieren, sind solche Sequenzen, welche trotz abweichender Nukleotidsequenz noch die gewünschten Funktionen besitzen. Funktionelle Äquivalente umfassen somit natürlich vorkommende Varianten der hierin beschriebenen Sequenzen sowie künstliche, z.B. durch chemische Synthese erhaltene, an den Kodon-Gebrauch einer Pflanze angepaßte, künstliche Nukleotid-Sequenzen.Functionally equivalent sequences which code for a DHO gene are those sequences which, despite a different nucleotide sequence, still have the desired functions. Functional equivalents thus include naturally occurring variants of the sequences described herein as well as artificial, e.g. Artificial nucleotide sequences obtained by chemical synthesis and adapted to the codon use of a plant.
Unter einem funktioneilen Äquivalent versteht man insbesondere auch natürliche oder künstliche Mutationen einer ursprünglich isolierten für eine DHO kodierende Sequenz, welche weiterhin die gewünschte Funktion zeigen. Mutationen umfassen Substitutionen, Additionen, Deletionen, Vertauschungen oder Insertionen eines oder mehrerer Nukleotidreste. Somit werden beispielsweise auch solche Nukleotidsequenzen durch die vorliegende Erfindung mit umfaßt, welche man durch Modifikation der DHO-Nukleotidsequenz erhält. Ziel einer solchen Modifikation kann z.B. die weitere Eingrenzung der darin enthaltenen kodierenden Sequenz oder z.B. auch die Einfügung weiterer Restriktionsenzym-Schnittstellen sein. Funktionelle Äquivalente sind auch solche Varianten, deren Funktion, verglichen mit dem Ausgangsgen bzw. Genfragment, abgeschwächt oder verstärkt ist.A functional equivalent is understood to mean, in particular, natural or artificial mutations of an originally isolated sequence coding for a DHO, which furthermore show the desired function. Mutations include substitutions, additions, deletions, exchanges or insertions of one or more nucleotide residues. Thus, for example, the present invention also encompasses those nucleotide sequences which are obtained by modifying the DHO nucleotide sequence. The aim of such a modification can, for example, be to further narrow down the coding sequence contained therein or, for example, also to insert further restriction enzyme interfaces. Functional equivalents are also those variants whose function is weakened or enhanced compared to the original gene or gene fragment.
5 Außerdem sind artifizielle DNA-Sequenzen geeignet, solange sie, wie oben beschrieben, die gewünschte Eigenschaft beispielsweise der Erhöhung des Stärke-Gehaltes in der Pflanze durch Über- expression des DHO-Gens in Kulturpflanzen vermitteln. Solche ar- tifiziellen DNA-Sequenzen können beispielsweise durch Ruckuber-5 In addition, artificial DNA sequences are suitable as long as, as described above, they impart the desired property, for example increasing the starch content in the plant by overexpressing the DHO gene in crop plants. Such artificial DNA sequences can be, for example, by Ruckuber
10 setzung mittels Molecular Modelling konstruierter Proteine, die DHO-Aktivitat aufweisen oder durch in vitro-Selektion ermittelt werden. Mögliche Techniken zur in vitro-Evolution von DNA zur Veränderung bzw. Verbesserung der DNA-Sequenzen sind beschrieben bei Patten, P.A. et al., Current Opinion in Biotechnology 8,10 Setting of proteins constructed by means of molecular modeling which have DHO activity or are determined by in vitro selection. Possible techniques for the in vitro evolution of DNA to change or improve the DNA sequences are described in Patten, P.A. et al., Current Opinion in Biotechnology 8,
15 724-733 ( 1997) oder bei Moore, J.C. et al . , Journal of Molecular Biology 272, 336 - 347 ( 1997). Besonders geeignet sind kodierende DNA-Sequenzen, die durch Rückübersetzung einer Polypeptidsequenz gemäß der f r die Wirtspflanze spezifischen Kodon-Nutzung erhalten wurden. Die spezifische Kodon-Nutzung kann ein mit pflanzen-15 724-733 (1997) or Moore, J.C. et al. , Journal of Molecular Biology 272, 336-347 (1997). Coding DNA sequences which are obtained by back-translating a polypeptide sequence according to the codon usage specific for the host plant are particularly suitable. The specific codon usage can be
20 genetischen Methoden vertrauter Fachmann durch Computerauswertungen anderer, bekannter Gene der zu transformierenden Pflanze leicht ermitteln.A person skilled in the art can easily determine 20 genetic methods by means of computer evaluations of other known genes of the plant to be transformed.
Als weitere geeignete äquivalente Nukleinsäure-Sequenzen sind zu 25 nennen Sequenzen, welche für Fusionsproteine kodieren, wobei Bestandteil des Fusionsproteins ein DHO-Polypeptid oder ein funk- tionell äquivalenter Teil davon ist. Der zweite Teil des Fusionsproteins kann z.B. ein weiteres Polypeptid mit enzymatischer Aktivität sein oder eine antigene Polypeptidsequenz mit deren 30 Hilfe ein Nachweis auf DHO-Expression möglich ist (z.B. myc-tag oder his-tag) . Bevorzugt handelt es sich dabei jedoch um eine regulative Proteinsequenz, wie z.B. ein Signal- oder Transitpeptid, das das DHO-Protein an den gewünschten Wirkort leitet.Other suitable equivalent nucleic acid sequences include 25 sequences which code for fusion proteins, part of the fusion protein being a DHO polypeptide or a functionally equivalent part thereof. The second part of the fusion protein can e.g. be another polypeptide with enzymatic activity or an antigenic polypeptide sequence that can be used to detect DHO expression (e.g. myc-tag or his-tag). However, this is preferably a regulatory protein sequence, such as e.g. a signal or transit peptide that directs the DHO protein to the desired site of action.
35 Erhöhung des Polysaccharidgehaltes bedeutet im Rahmen der vorliegenden Erfindung beispielsweise die künstlich erworbene Fähigkeit einer erhöhten Stärke-Biosyntheseleistung durch funktioneile Überexpression des DHO-Gens in der Pflanze gegenüber der nicht gentechnisch modifizierten Pflanze für die Dauer mindestens einerIn the context of the present invention, increasing the polysaccharide content means, for example, the artificially acquired ability of an increased starch biosynthesis performance by functional overexpression of the DHO gene in the plant compared to the non-genetically modified plant for at least one period
40 Pflanzengeneration.40 generation of plants.
Der Biosyntheseort von Starke beispielsweise ist im allgemeinen das Blattgewebe, so daß eine blattspezifische Expression des DHO- Gens sinnvoll ist. Es ist jedoch naheliegend, daß die Stärkebio- 45 Synthese nicht auf das Blattgewebe beschränkt sein muß, sondern auch in allen übrigen Teilen der Pflanze - beispielsweise in fetthaltigen Samen oder in den Knollen - gewebespezifisch erfolgen kann .Starke's biosynthesis site, for example, is generally leaf tissue, so that leaf-specific expression of the DHO gene makes sense. However, it is obvious that the starch bio-synthesis must not be restricted to the leaf tissue, but also in all other parts of the plant - for example in fatty seeds or in the tubers - tissue-specific.
Darüberhinaus ist eine konstitutive Expression des exogenen DHO- Gens von Vorteil. Andererseits kann aber auch eine induzierbare Expression wünschenswert erscheinen.In addition, constitutive expression of the exogenous DHO gene is advantageous. On the other hand, inducible expression may also appear desirable.
Die Wirksamkeit der Expression des transgen exprimierten DHO-Gens kann beispielsweise in vi tro durch Sproßmeristemvermehrung ermit- telt werden. Zudem kann eine in Art und Höhe veränderte Expression des DHO-Gens und deren Auswirkung auf die Polysaccharid-Bio- syntheseleistung an Testpflanzen in Gewächshausversuchen getestet werden.The effectiveness of the expression of the transgenically expressed DHO gene can be determined, for example, in vitro by propagation of the shoot meristem. In addition, a change in the type and level of expression of the DHO gene and its effect on the polysaccharide biosynthesis performance on test plants can be tested in greenhouse experiments.
Gegenstand der Erfindung sind außerdem transgene Pflanzen, transformiert mit einer Expressionskassette enthaltend eine DHO-Gense- quenz oder mit dieser hybridisierende DNA-Sequenzen, sowie transgene Zellen, Gewebe, Teile und Vermehrungsgut solcher Pflanzen. Besonders bevorzugt sind dabei transgene Kulturp lanzen, wie z.B. Gerste, Weizen, Roggen, Hafer, Mais, Soja, Reis, Baumwolle, Zuckerrübe, Canola, Sonnenblume, Flachs, Hanf, Kartoffel, Tabak, Tomate, Raps, Tapioka, Maniok, Pfeilwurz, Alfalfa, Salat und die verschiedenen Baum-, Nuß- und Weinspezies.The invention also relates to transgenic plants transformed with an expression cassette containing a DHO gene sequence or DNA sequences hybridizing therewith, and transgenic cells, tissues, parts and propagation material of such plants. Transgenic crop plants, such as e.g. Barley, wheat, rye, oats, corn, soybeans, rice, cotton, sugar beet, canola, sunflower, flax, hemp, potato, tobacco, tomato, rapeseed, tapioca, cassava, arrowroot, alfalfa, lettuce and the various tree nuts - and wine species.
Pflanzen im Sinne der Erfindung sind mono- und dikotyle Pflanzen oder Algen.Plants in the sense of the invention are mono- and dicotyledonous plants or algae.
Weitere Gegenstände der Erfindung sind:Further objects of the invention are:
- Verfahren zur Transformation einer Pflanze dadurch gekennzeichnet, daß man Expressionskassetten enthaltend eine DHO- Gensequenz oder mit dieser hybridisierende DNA-Sequenzen in eine Pflanzenzelle, in Kallusgewebe, eine ganze Pflanze oder Protoplasten von Pflanzen einbringt.- Process for transforming a plant, characterized in that expression cassettes containing a DHO gene sequence or DNA sequences hybridizing therewith are introduced into a plant cell, into callus tissue, an entire plant or protoplasts of plants.
Verwendung einer DHO-DNA-Gensequenz oder mit dieser hybridisierende DNA-Sequenzen zur Herstellung von Pflanzen mit erhöhtem Polysaccharidgehalt durch Expression dieser DHO-DNA- Sequenz in Pflanzen.Use of a DHO DNA gene sequence or DNA sequences hybridizing therewith for the production of plants with an increased polysaccharide content by expression of this DHO DNA sequence in plants.
Die Erfindung wird durch die nun folgenden Beispiele erläutert, ist aber nicht auf diese beschränkt:The invention is illustrated by the following examples, but is not limited to these:
Allgemeine Klonierungsverfahren: Klonierungsverfahren wie z.B. Restriktionsspaltungen, Agarose- Gelelektrophorese, Reinigung von DNA-Fragmenten, Transfer von Nukleinsäuren auf Nitrozellulose und Nylon Membranen, Verknüpfen von DNA-Fragmenten, Transformation von Escherichia coli Zellen, Anzucht von Bakterien und Sequenzanalyse rekombinanter DNA wurden wie bei Sambrook et al. (1989) (Cold Spring Harbor Laboratory Press: ISBN 0-87969-309-6) beschrieben durchgeführt.General cloning procedures: Cloning methods such as restriction cleavage, agarose gel electrophoresis, purification of DNA fragments, transfer of nucleic acids to nitrocellulose and nylon membranes, linking of DNA fragments, transformation of Escherichia coli cells, cultivation of bacteria and sequence analysis of recombinant DNA were carried out as in Sambrook et al. (1989) (Cold Spring Harbor Laboratory Press: ISBN 0-87969-309-6).
Sequenzanalyse rekombinanter DNA:Sequence analysis of recombinant DNA:
Die Sequenzierung rekombinanter DNA-Moleküle erfolgte mit einem Laserfluoreszenz-DNA-Sequenzierer der Firma ABI nach der Methode von Sanger (Sanger et al. (1977) Proc. Natl. Acad. Sei. USA74, 5463-5467) . Fragmente resultierend aus einer Polymerase Kettenre- aktion wurden zur Vermeidung von Polymerasefehlern in zu expri- ierenden Konstrukten sequenziert und überprüft.The sequencing of recombinant DNA molecules was carried out with a laser fluorescence DNA sequencer from ABI according to the method of Sanger (Sanger et al. (1977) Proc. Natl. Acad. Sci. USA74, 5463-5467). Fragments resulting from a polymerase chain reaction were sequenced and checked in order to avoid polymerase errors in constructs to be expressed.
Gesamt-RNA aus pflanzlichen Geweben wurde wie bei Logemann et al . (1987, Anal. Biochem. 163, 21) isoliert. Für die Analyse wurden jeweils 20 μg RNA in einem Formaldehyd-haltigen l,5%igen Agarose - gel aufgetrennt und auf Nylon Membranen (Hybond, Amersham) überführt. Der Nachweis spezifischer Transkripte wurde wie bei Ama- sino beschrieben durchgeführt (1986, Anal. Biochem. 152, 304). Die als Sonde eingesetzten cDNA-Fragmente wurden mit einem Random Primed DNA Labeling Kit (Boehringer, Mannheim) radioaktiv markiert und nach Standardmethoden hybrisiert (Siehe Hybon -Benut - zerhinweise, Amersham) . HyridisierungsSignale wurden durch Auto- radiographie mithilfe von X-OMAT AR Filmen der Fa. Kodak sichtbar gemacht.Total RNA from plant tissues was, as in Logemann et al. (1987, Anal. Biochem. 163, 21). For the analysis, 20 μg RNA was separated in a 1.5% agarose gel containing formaldehyde and transferred to nylon membranes (Hybond, Amersham). The detection of specific transcripts was carried out as described in Aminos (1986, Anal. Biochem. 152, 304). The cDNA fragments used as a probe were radioactively labeled with a random primed DNA labeling kit (Boehringer, Mannheim) and hybridized according to standard methods (see Hybon user references, Amersham). Hyridization signals were visualized by autoradiography using X-OMAT AR films from Kodak.
Beispiel 1example 1
Erhöhung der Pyrimidin-Nukleotidkonzentration in Kartoffelknollenscheiben durch Fütterung mit Orotat oder Uridin.Increase in pyrimidine nucleotide concentration in potato tuber slices by feeding with orotate or uridine.
Kartoffelpflanzen (Solanum tuberosu L. cv. Desiree, Saatzucht Fritz Lange, Bad Schwartau) wurden in Wachstumskammern (Bestrahlungsstärke: 350 μmol Photonen ιrτ s-1, 14 h /10 h Tag- / Nacht- Rhythmus, Temperatur: 20 °C, 50 % relative Luftfeuchte) in 3 1 Töpfen auf Erde (mit 100 g "Hakaphos grün" [BASF-AG,Potato plants (Solanum tuberosu L. cv.Desiree, Saatzucht Fritz Lange, Bad Schwartau) were grown in growth chambers (irradiance: 350 μmol photons ιrτ s -1 , 14 h / 10 h day / night rhythm, temperature: 20 ° C, 50 % relative humidity) in 3 1 pots on earth (with 100 g "Hakaphos green" [BASF-AG,
Ludwigshafen] pro 230 1) oder im Gewächshaus mit Zusatzlicht (150 μmol Photonen m-2s-1) angezogen. Wachsende Knollen von täglich gewässerten Pflanzen wurden für die Experimente eingesetzt.Ludwigshafen] per 230 1) or in a greenhouse with additional light (150 μmol photons m- 2 s -1 ). Growing tubers from plants watered daily were used for the experiments.
Knollenscheiben von 2 mm Dicke und 8 mm Durchmesser (ca. 0,1 g) wurden präpariert, wie in Geigenberger et al . (1997, Planta 201, 502-518) beschrieben. Nach dreimaligem Waschen mit 10 mM 2- (N- morpholino) -Ethan-Sulfonsäure (Mes) (pH 6.5; KOH) wurden die Scheiben in 100 ml Erlenmeyerkolben bei 90 upm im entsprechenden Medium (8 Scheiben in 4 ml) inkubiert. Nach 90 Minuten wurden [U-1 C] -Glucose oder fU-1 C] -Sucrose (1,1 kBq μmol-1; Amersham- Buchler) zugegeben und weitere 2 h inkubiert. Die Scheiben wurden 3x in Puffer gewaschen und in flussigem Stickstoff schockgefroren.Tuber disks with a thickness of 2 mm and a diameter of 8 mm (approx. 0.1 g) were prepared as described in Geigenberger et al. (1997, Planta 201, 502-518). After washing three times with 10 mM 2- (N- morpholino) -ethane-sulfonic acid (Mes) (pH 6.5; KOH) the disks were incubated in 100 ml Erlenmeyer flasks at 90 rpm in the appropriate medium (8 disks in 4 ml). After 90 minutes, [U- 1 C] glucose or fU- 1 C] sucrose (1.1 kBq μmol- 1 ; Amersham-Buchler) were added and incubated for a further 2 h. The slices were washed 3 times in buffer and snap frozen in liquid nitrogen.
Die Knollenscheiben wurden mit 80 % (v/v) Ethanol extrahiert (1 ml für 2 Scheiben) und in drei Folgeschritten reextrahiert (80 % (v/v) Ethanol, 50 % (v/v) Ethanol, H20) . Die kombinierten Überstände wurden bei 47 °C im Luftstrom getrocknet und in 1 ml H0 aufgenommen. Diese losliche Fraktion wurde wie bei Quick et al . (1989, Planta 177, 536-546) durch Ionenaustauschchromatographie in neutrale, basische und saure Fraktionen getrennt. Die neutrale Fraktion wurde nach Gefriertrocknung in 100 μl H20 aufgenommen und mittels Dunnschichtchromatographie analysiert (Geigenberger et al . 1997, Planta 201, 502-518). Zur Messung der Phosphatester wurden 150 μl der loslichen Fraktion in 50 μl Puffer (10 mM Mes, pH 6.5; KOH) mit oder ohne 1 U saurer Phosphatase aus Kartoffel (grade II, Boehringer, Mannheim) für 3 h bei 37 °C inkubiert und nach 2-minutigem Kochen durch Ionenaustauschchromatographie analysiert (Geigenberger et al . , 1997). Nukleotidkonzentrationen wurden wie bei Geigenberger et al. beschrieben aus Trichloressig- Säureextrakten durch HPLC-Analyse mittels einer Partisil-SAX An- ionenaustauschersäule bestimmt. Aus dem unlöslichen Ruckstand nach der Ethanolextraktion wurden Stärke, Protein und Zellwandkomponenten wie bei Merlo et al. (1993, J. Plant Physiol. 142: 392-402) beschrieben bestimmt.The tuber slices were extracted with 80% (v / v) ethanol (1 ml for 2 slices) and re-extracted in three subsequent steps (80% (v / v) ethanol, 50% (v / v) ethanol, H 2 0). The combined supernatants were dried in a stream of air at 47 ° C. and taken up in 1 ml of H0. This soluble fraction was, as in Quick et al. (1989, Planta 177, 536-546) separated into neutral, basic and acidic fractions by ion exchange chromatography. After freeze-drying, the neutral fraction was taken up in 100 μl of H 2 O and analyzed by means of thin layer chromatography (Geigenberger et al. 1997, Planta 201, 502-518). To measure the phosphate esters, 150 μl of the soluble fraction in 50 μl buffer (10 mM Mes, pH 6.5; KOH) with or without 1 U acidic phosphatase from potato (grade II, Boehringer, Mannheim) were incubated for 3 h at 37 ° C. and after boiling for 2 minutes, analyzed by ion exchange chromatography (Geigenberger et al., 1997). Nucleotide concentrations were determined as in Geigenberger et al. described from trichloroacetic acid extracts determined by HPLC analysis using a Partisil-SAX anion exchange column. From the insoluble residue after ethanol extraction, starch, protein and cell wall components as in Merlo et al. (1993, J. Plant Physiol. 142: 392-402).
Orotat und Uridm sind Vorstufen der Uridinnukleotide. Im Folgenden sollte die Frage untersucht werden, ob eine Futterung mit Orotat oder Uridin einen Einfluß auf den Nucleotidgehalt in Knollenscheiben hat. Hierzu wurden Knollenscheiben 10 Wochen alter Kartoffelpflanzen für 3 Stunden in Gegenwart von 1 mM Glucose und der entsprechenden Uridinnukleotidvorstufen inkubiert. Anschließend wurden die Nukleotidgehalte gemessen.Orotate and Uridm are precursors to uridine nucleotides. In the following, the question should be examined whether feeding with orotate or uridine has an influence on the nucleotide content in tuber disks. For this purpose, tuber slices of 10-week-old potato plants were incubated for 3 hours in the presence of 1 mM glucose and the corresponding uridine nucleotide precursors. The nucleotide contents were then measured.
Abbildung 2 zeigt die Nukleotidkonzentration in frisch präparier- ten Kartoffelknollenscheiben von wachsenden Knollen 10 Wochen alter Pflanzen ohne bzw. mit Futterung verschiedener Nukleotidvor- stufen ( Inkubation f r 3 Stunden in Gegenwart von 10 mM Mes-KOH (ph 6,5), 300 mM Mannitol und 1 mM Glucose. Im Vergleich zu nicht inkubierten Proben war eine Abnahme des Gesamtgehaltes an Uridin- nukleotiden (UDPGlc + UTP + UDP; UMP war vernachlässigbar) um 30 - 40 % nach Inkubation mit 1 mM Glucose festzustellen (Abb. 2A) . Eine Inkubation im gleichen Puffer, zusätzlich enthaltend 10 mM Uridin oder 10 mM Orotat verhinderte den Effekt und führte darüber hinaus zu einer Zunahme des Gesamtgehaltes an Uridinnukleo- tiden von 15 - 25 %, die auf alle untersuchten Uridinnukleotide zurück ging (Abb. 2B, C, D) . Dabei war in allen Experimenten die Erhöhung durch Gabe von Orotat geringfügig größer als durch Gabe von Uridin. Inkubation mit geringeren Konzentrationen an Orotat oder Uridin (5 mM) führten zu einer geringeren Erhöhung der gesamt Uridinnukleotidkonzentration (nicht abgebildet) .Figure 2 shows the nucleotide concentration in freshly prepared potato tuber slices from growing tubers of 10-week-old plants with or without feeding various nucleotide precursors (incubation for 3 hours in the presence of 10 mM Mes-KOH (pH 6.5), 300 mM mannitol) and 1 mM glucose Compared to non-incubated samples, the total uridine nucleotide content (UDPGlc + UTP + UDP; UMP was negligible) decreased by 30-40% after incubation with 1 mM glucose (Fig. 2A) Incubation in the same buffer, additionally containing 10 mM Uridine or 10 mM orotate prevented the effect and moreover led to an increase in the total uridine nucleotide content of 15-25%, which was attributable to all uridine nucleotides examined (Fig. 2B, C, D). In all experiments, the increase by administration of orotate was slightly larger than by administration of uridine. Incubation with lower concentrations of orotate or uridine (5 mM) resulted in a smaller increase in the total uridine nucleotide concentration (not shown).
Im Gegensatz zum Uridinnukleotidpool stieg die Konzentration an Adenylaten und Guanylaten (Abb. 2E,F) in kontrollinkubierten Scheiben (ohne Uridin bzw. Orotat) im Vergleich zu nicht inkubierten Scheiben leicht an. Diese Zunahme war unabhängig von einer Inkubation mit Orotat oder Uridin, was im Einklang mit der Annahme steht, daß Orotat und Uridin spezifische Vorstufen für die Uridinnukleotide und nicht für die Purinnukleotide darstellen. Andererseits bewirkte eine Inkubation mit 5 mM Adenin keine Erhöhung der Uridinnukleotide aber eine etwa 2fache Erhöhung der Gesamt-Adenylate und -Guanylate (Abb. 2E, F) .In contrast to the uridine nucleotide pool, the concentration of adenylates and guanylates (Fig. 2E, F) in control-incubated disks (without uridine or orotate) increased slightly compared to non-incubated disks. This increase was independent of incubation with orotate or uridine, which is consistent with the assumption that orotate and uridine are specific precursors for the uridine nucleotides and not for the purine nucleotides. On the other hand, incubation with 5 mM adenine did not cause an increase in the uridine nucleotides but an approximately 2-fold increase in the total adenylates and guanylates (Fig. 2E, F).
Diese Ergebnisse zeigen, daß es möglich ist, durch Fütterung von Uridinnukleotidvorstufen wie Orotat oder Uridin die Konzentration von Uridinnukleotiden in Pflanzenzellen zu erhöhen.These results show that it is possible to increase the concentration of uridine nucleotides in plant cells by feeding uridine nucleotide precursors such as orotate or uridine.
Beispiel 2Example 2
Erhöhung der Stärkesynthese in Kartoffelknollenscheiben durch Fütterung mit Orotat oder Uridin.Increase in starch synthesis in potato tuber slices by feeding with orotate or uridine.
Um die Frage zu beantworten, ob erhöhte Konzentrationen an Uridinnukleotiden einen Effekt auf den Sucroseabbau und den Stärkegehalt haben, wurden Knollenscheiben mit 100 mM 14C-Sucrose in Gegenwart sowie in Abwesenheit von 10 mM Orotat inkubiert. Ebenso wie in Gegenwart von Glucose führte die Fütterung mit Orotat zur Erhöhung der Uridinnukleotidkonzentrationen, ohne die Adenylat- und Guanylatkonzentrationen zu beeinflussen. Abb. 3 zeigt die Nu- kleotidkonzentration in frisch präparierten Kartoffelscheiben von wachsenden Knollen 10 Wochen alter Pflanzen ohne bzw. mit Fütterung von 10 mM Orotat ( Inkubation für 3 Stunden in Gegenwart von 10 mM Mes-KOH ( pH6,5) und 100 mM Sucrose. Abbildung 4 zeigt die Metabolisierung von 1 C - Sucrose frisch präparierter Kartoffelknollenscheiben von wachsenden Knollen 10 Wochen alter Pflanzen ohne bzw. mit Fütterung von 10 mM Orotat ( Inkubation für 90 Minuten in Gegenwart von 100 mM Sucrose. Anschließende Zugabe von 1 C - Sucrose ( 1,1 kBq μmol"1) und Inkubation für weitere 2 Stunden) . Orotat führte zu einer geringfügigen Steigerung der Aufnahme von 14C-Sucrose (Abb. 4A) sowie einer 2-fachen Zunahme der 14C-Sucrose Degradation (von 8 % der absorbierten Radioaktivität in Abwesenheit von Orotat auf 15 % - 18 % in Anwesenheit von Orotat, Abb. 4B) . Die Orotatfütterung führte zu einer Steigerung des Einbaus an Radioaktivität in Stärke (Abb. 4C) , sowie einer Ab- nähme des Einbaus in Phosphatester (Abb. 4E) , organische Säuren (Abb. 4F) und freie Aminosäuren (Abb 4G) . Der Einbau in Zellwandkomponenten und Proteine blieb im Wesentlichen unverändert (Abb. 4D, H) .To answer the question whether increased concentrations of uridine nucleotides have an effect on sucrose breakdown and starch content, tuber slices were incubated with 100 mM 14 C-sucrose in the presence and in the absence of 10 mM orotate. As in the presence of glucose, feeding with orotate led to an increase in uridine nucleotide concentrations without affecting adenylate and guanylate concentrations. Fig. 3 shows the nucleotide concentration in freshly prepared potato slices of growing tubers of 10-week-old plants without or with feeding 10 mM orotate (incubation for 3 hours in the presence of 10 mM Mes-KOH (pH 6.5) and 100 mM sucrose) Figure 4 shows the metabolism of 1 C - sucrose freshly prepared potato tuber slices from growing tubers of plants 10 weeks old without or with feeding of 10 mM orotate (incubation for 90 minutes in the presence of 100 mM sucrose. Subsequent addition of 1 C - sucrose ( 1.1 kBq μmol " 1 ) and incubation for a further 2 hours). Orotat led to a slight increase in the intake of 14 C-sucrose (Fig. 4A) and a 2-fold increase in the 14 C-sucrose degradation (from 8% of the absorbed radioactivity in the absence of orotate to 15% - 18% in the presence of orotate, Fig. 4B). Orotate feeding led to an increase in the incorporation of radioactivity in starch (Fig. 4C), as well as a decrease in incorporation in phosphate ester (Fig. 4E), organic acids (Fig. 4F) and free amino acids (Fig. 4G). The incorporation into cell wall components and proteins remained essentially unchanged (Fig. 4D, H).
Orotat führte insgesamt zu einer 2,4-fachen Steigerung des absoluten Flux an Sucrose in Stärke (Tab. 1) .Overall, orotate led to a 2.4-fold increase in the absolute flux of sucrose in starch (Table 1).
Berechnung der absoluten Stärkesyntheserate in An- und Abwesenheit von Orotat über die spezifische Aktivität des Hexosephosp- hat-Pools. Die spezifische Aktivität wurde errechnet, indem die gemessene Radioaktivität in Phosphatestern durch die Summe des Kohlenstoffs im Hexosephosphat-Pool (Glucose-6-Phosphat + Fruc- tose-6-Phosphat + Glucose-1-Phosphat ; Daten nicht gezeigt) geteilt wurde. Mittelwerte +/- Standardabweichung (n = 4)Calculation of the absolute starch synthesis rate in the presence and absence of orotate via the specific activity of the hexose phosphate pool. The specific activity was calculated by dividing the measured radioactivity in phosphate esters by the sum of the carbon in the hexosephosphate pool (glucose-6-phosphate + fructose-6-phosphate + glucose-1-phosphate; data not shown). Mean +/- standard deviation (n = 4)
Tabelle 1Table 1
0 Beispiel 30 Example 3
Erzeugung transgener TabakpflanzenGeneration of transgenic tobacco plants
Zur Erzeugung transgener Tabakpflanzen wurden binäre Vektoren in 5 Agrobacterium tumefaciens C58Cl:pGV2260 transformiert (Deblaere et al, 1984, Nucl. Acids . Res . 13, 4777-4788). Zur Transformation von Tabakpflanzen (Nicotiana tabacum cv. Samsun NN) , wurde eine 1:50 Verdünnung einer Übernachtkultur einer positiv transformierten Agrobakterienkolonie in Murashige-Skoog Medium (Murashige und 0 Skoog 1962 Physiol. Plant. 15, 473) mit 2% Saccharose (2MS-Me- dium) benutzt. Blattscheiben steriler Pflanzen (zu je ca. 1 cm2) wurden in einer Petrischale mit einer 1:50 Agrobakterienverdün- nung für 5-10 Minuten inkubiert. Es folgte eine 2-tägige Inkubation in Dunkelheit bei 25°C auf 2MS-Medium mit 0,8% Bacto-Agar. e Die Kultivierung wurde nach 2 Tagen mit 16 Stunden Licht / 8 Stunden Dunkelheit weitergeführt und in wöchentlichem Rhythmus auf MS-Medium mit 500 mg/1 Claforan (Cefotaxime-Natrium) , 50 mg/1 Kanamycin, 1 mg/1 Benzylaminopurin (BAP) , 0,2 mg/1 Naphtylessig- säure und 1,6 g/1 Glukose weitergeführt. Wachsende Sprosse wurden auf MS-Medium mit 2% Saccharose, 250 mg/1 Claforan und 0,8% Bacto-Agar überführt.To generate transgenic tobacco plants, binary vectors were transformed into 5 Agrobacterium tumefaciens C58Cl: pGV2260 (Deblaere et al, 1984, Nucl. Acids. Res. 13, 4777-4788). For the transformation of tobacco plants (Nicotiana tabacum cv. Samsun NN), a 1:50 dilution of an overnight culture of a positively transformed agrobacterial colony in Murashige-Skoog medium (Murashige and 0 Skoog 1962 Physiol. Plant. 15, 473) with 2% sucrose (2MS -Medium) is used. Leaf disks of sterile plants (each about 1 cm 2 ) were incubated in a Petri dish with a 1:50 agrobacterial dilution for 5-10 minutes. This was followed by a 2-day incubation in the dark at 25 ° C. on 2MS medium with 0.8% Bacto agar. e The cultivation was continued after 2 days with 16 hours of light / 8 hours of darkness and in a weekly rhythm on MS medium with 500 mg / 1 claforan (cefotaxime sodium), 50 mg / 1 Kanamycin, 1 mg / 1 benzylaminopurine (BAP), 0.2 mg / 1 naphthylacetic acid and 1.6 g / 1 glucose. Growing shoots were transferred to MS medium with 2% sucrose, 250 mg / 1 Claforan and 0.8% Bacto agar.
Beispiel 4Example 4
Sequenzanalyse der cDNA Klone codierend für ein Protein mit Dihydroorotase Aktivität.Sequence analysis of the cDNA clones coding for a protein with dihydroorotase activity.
Die resultierenden 36 cDNA Klone codieren für ein Polypeptid mit Homologie zu Dihydroorotasen aus anderen Organismen. Die Homologie wurde mit dem Programm BLASTP erhalten. (Altschul et al., Nucleic Acids Res . (1997) 25, 3389-3402). Demnach ist das Protein zu 78 % identisch zur Dihydroorotase aus Arabidopsis thaliana,The resulting 36 cDNA clones code for a polypeptide with homology to dihydroorotases from other organisms. The homology was obtained with the BLASTP program. (Altschul et al., Nucleic Acids Res. (1997) 25, 3389-3402). Accordingly, the protein is 78% identical to Arabidopsis thaliana dihydroorotase,
58 % zu Synechocystis, 55% zu E. coli und Pseudomonas putida. Der längste Klon wurde pyrCSt5 genannt. Das Plasmid beträgt die Bezeichnung pBSSK-pyrCSt5. Die cDNA (siehe SEQ-ID No. 1) hat einen offenen Leseraster von 1046 Basenpaaren mit einem Stop- Codon in Position 1047-1049. Die Aminosäuresequenz beginnt mit der dritten Base im Leseraster und kann in ein 348 Aminosäuren langes Polypeptid übersetzt werden (siehe SEQ-ID No. 2). Dies entspricht der Länge prokaryotischer Dihydroorotase-codierender Sequenzen.58% to Synechocystis, 55% to E. coli and Pseudomonas putida. The longest clone was called pyrCSt5. The plasmid is called pBSSK-pyrCSt5. The cDNA (see SEQ ID No. 1) has an open reading frame of 1046 base pairs with a stop codon in position 1047-1049. The amino acid sequence begins with the third base in the reading frame and can be translated into a 348 amino acid polypeptide (see SEQ-ID No. 2). This corresponds to the length of prokaryotic dihydroorotase coding sequences.
Beispiel 5Example 5
Isolation einer cDNA codierend für eine funktioneile pflanzliche DihydroorotaseIsolation of a cDNA coding for a functional plant dihydroorotase
Ein Klon codierend für Dihydroorotase wurde aus Kartoffel über funktioneile Komplementation einer E.coli Mutante erhalten. Es wurde die Mutante CGSC5152 (CS101-2U5) des E. coli Genetic Stock Centers verwendet, die eine Mutation im pyrC Genlokus codierend für eine Dihydroorotase trägt. Die Komplementation erfolgte durch Elektrotransformation kompetenter Zellen des Stammes CGSC5152 mit einer cDNA Bank in dem Vektorplasmid pBS SK- . Die zugrunde liegende Lambda ZAPII Bank (Stratagene) wurde nach Standardvorschriften ungerichtet mit EcoRI/Notl Linkern kloniert. Die RNA- Matrize für die cDNA wurde aus sink leaves von Kartoffel (kleiner 1 cm Blättchen von 10 Wochen alten Kartoffelpflanzen geerntet im Gewaechshaus gezogen) isoliert.A clone coding for dihydroorotase was obtained from potato via the functional complementation of an E. coli mutant. The mutant CGSC5152 (CS101-2U5) of the E. coli Genetic Stock Center was used, which carries a mutation in the pyrC gene locus coding for a dihydroorotase. The complementation was carried out by electrotransformation of competent cells of the CGSC5152 strain with a cDNA bank in the vector plasmid pBS SK-. The underlying Lambda ZAPII bank (Stratagene) was cloned undirected using EcoRI / Notl linkers according to standard regulations. The RNA template for the cDNA was isolated from sink leaves of potato (small 1 cm leaflet from 10 week old potato plants harvested in a greenhouse).
Die transformierten E. coli Zellen wurden auf Minimalmedium M9 plattiert (Sambrook et al . , 1989 s.o.), das zusätzlich Methionin (20 mg/1), Ampicillin (100 mg/1) und IPTG (2.5 mM) enthielt. Es wurden insgesamt 4 Microgramm der Bank in 8 Ansätzen transfor- miert und es konnten 36 Klone erhalten werden, die sich nachThe transformed E. coli cells were plated on minimal medium M9 (Sambrook et al., 1989 see above), which additionally contained methionine (20 mg / 1), ampicillin (100 mg / 1) and IPTG (2.5 mM). A total of 4 micrograms of the bank were transformed in 8 approaches miert and 36 clones could be obtained, which follow
Untersuchung durch Restriktionsspaltung als gleich erwiesen.Examination by restriction cleavage proved to be the same.
Beispiel 6Example 6
Erzeugung transgener Pflanzen, welche ein Enzym mit Dihydroorota- se-Aktivität aus Kartoffel überexprimieren.Generation of transgenic plants which overexpress an enzyme with dihydroorotase activity from potatoes.
Es wurde eine cDNA hergestellt, die für ein Enzym mit Dihydrooro- tase-Aktivität aus Kartoffel codiert, das an eine zum Import des Proteins in die Plastiden führende Signalsequenz (entnommen einem Enzym mit Tranketolase-Aktivität aus Tabak) fusioniert wurde. Hierzu wurden zunächst anhand der pBSSK-pyrCSt5 cDNA die Oligo- nukleotide 5' -GTCGACATGGAGCTCTCAATCACACAACC-3 ' undA cDNA was produced which codes for an enzyme with dihydroorotase activity from potato which was fused to a signal sequence leading to the import of the protein into the plastids (taken from an enzyme with tranketolase activity from tobacco). For this purpose, the oligonucleotides 5 '-GTCGACATGGAGCTCTCAATCACACAACC-3' and... Were first of all determined using the pBSSK-pyrCSt5 cDNA
5'-GTCGACACACCTACAGTCTATATCTTTGG-3' für eine Polymerase Ketten - reakion (PCR) abgeleitet. Durch eine PCR mit pBSSK-pyrCSt5 als Matrize wurden Sall-Restriktionsschnittstellen vor dem Startcodon sowie nach dem Stopcodon der Dihydroorotase cDNA eingeführt. Die Reaktionsgemische enthielten ca. 1 ng/microl Matrizen DNA, 0,5 microM der Oligonukleotide und, 200 microM Desoxy-Nukleotide (Pharmacia), 50 mM KC1, 10 mM Tris-HCl (pH 8,3 bei 25 °C, 1 , 5 mM MgCl2) und 0.02 U/microl Pwo Polymerase (Boehringer Mannheim) und wurden in einer PCR-Maschine der Firma Perkin Eimer mit folgendem Temperaturprogramm inkubiert:5'-GTCGACACACCTACAGTCTATATCTTTGG-3 'derived for a polymerase chain reaction (PCR). PCR with pBSSK-pyrCSt5 as a template introduced Sall restriction sites before the start codon and after the stop codon of the dihydroorotase cDNA. The reaction mixtures contained approx. 1 ng / microl template DNA, 0.5 microM of the oligonucleotides and, 200 microM deoxy nucleotides (Pharmacia), 50 mM KC1, 10 mM Tris-HCl (pH 8.3 at 25 ° C., 1.1 5 mM MgCl 2 ) and 0.02 U / microl Pwo polymerase (Boehringer Mannheim) and were incubated in a PCR machine from Perkin Elmer with the following temperature program:
Anlagerungstemperatur: 50°C, 45 secAnnealing temperature: 50 ° C, 45 sec
Denaturierungstemperatur : 95°C, 45 sec Elongationstemperatur : 72°C, 120 secDenaturation temperature: 95 ° C, 45 sec. Elongation temperature: 72 ° C, 120 sec
Anzahl der Zyklen: 30Number of cycles: 30
Das erhaltene Fragment von ca. 1,1 kbp wurde in den mit EcoRV gespaltenen Vektor pBluescript SK- (Stratagene) ligiert. Durch Kontrollspaltung wurde ein Klon identifiziert K4, dessen Insert durch Sall in voller Länge exzisierbar ist (1118 bp) . Das Insert K4 wurde vollständig sequenziert, um Polymerasefehler auszuschließen.The fragment of approximately 1.1 kbp obtained was ligated into the vector pBluescript SK- (Stratagene) which had been cleaved with EcoRV. A clone K4 was identified by control cleavage, the insert of which can be excised in full length by Sall (1118 bp). The insert K4 was completely sequenced to rule out polymerase errors.
Für die Transformation von Pflanzen wurde ein Transfervektor erzeugt, indem das 1118 bp Sall-Fragment aus K4 in den mit Sall gespaltenen Vektor pTK-TP-BinAR9 (R. Badur, 1998 Doktorarbeit, Universität Göttingen) ligiert wurde. Die Orientierung des In- serts wurde durch Spaltung mit Kpnl kontrolliert (es resultierte ein Fragtment von ca 980 bp) . Auf diese Weise wurde eine Fusion des Leserasters der Dihydroorotase aus Kartoffel an ein plastidä- res Transitpeptid, bestehend aus den N-terminalen 60 Aminosäuren der Transketolase aus Tabak (Genbank Acc. #CAA03393 ) erreicht (Konstrukt K5) . Die fusionierte cDNA Sequenz steht unter Kontrolle des Blumenkohlmosaik-Virus 35S-Promoters und des Octopin- synthase-Terminators aus Agrobacterium tumefaciens.A transfer vector was generated for the transformation of plants by ligating the 1118 bp Sall fragment from K4 into the vector pTK-TP-BinAR9 cleaved with Sall (R. Badur, 1998 doctoral thesis, University of Göttingen). The orientation of the insert was checked by cleavage with Kpnl (a fragment of approx. 980 bp resulted). In this way, the reading frame of the potato dihydroorotase was fused to a plastid transit peptide consisting of the N-terminal 60 amino acids achieved the tobacco transketolase (Genbank Acc. # CAA03393) (construct K5). The fused cDNA sequence is under the control of the cauliflower mosaic virus 35S promoter and the octopine synthase terminator from Agrobacterium tumefaciens.
Das Konstrukt K5 wurde zur Transformation von Tabak, Arabidopsis thaliana und Kartoffelpflanzen eingesetzt.The construct K5 was used to transform tobacco, Arabidopsis thaliana and potato plants.
Beispiel 7Example 7
Erzeugung transgener Arabidopsis thaliana PflanzenGeneration of transgenic Arabidopsis thaliana plants
Die Transformation von Arabidopsis thaliana erfolgte wie bei Bechtold, N. , Ellis, J. and Pelletier, G. in Planta, Agrobacte- rium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants, C. R. Acad. Sei. Paris, Life Sciences 316(1993), 1194 - 1199 beschrieben.Arabidopsis thaliana was transformed as in Bechtold, N., Ellis, J. and Pelletier, G. in Planta, Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants, C.R. Acad. Be. Paris, Life Sciences 316 (1993), 1194-1199.
Beispiel 8Example 8
Die Transformation von Kartoffelpflanzen (Solanum tuberosum, cv. Desiree) erfolgte wie bei Dietze et al. , in Gene Transfer to Plants, 1995, Potrykus und Spangenberg (Editoren), Springer, Berlin, beschrieben.The transformation of potato plants (Solanum tuberosum, cv. Desiree) was carried out as in Dietze et al. , in Gene Transfer to Plants, 1995, Potrykus and Spangenberg (editors), Springer, Berlin.
Beispiel 9Example 9
Die Transformation von Maispflanzen erfolgte wie bei Pareddy, D., Petolino, J. , Skokut, T., Hopkins, N. , Miller, M. , Welter, M. , Smith, K. , Clayton, D. , Pescitelli, S., Gould, A. , Maize Transformation via Helium Blasting. Maydica. 42(2): 143-154, 1997, beschrieben.The transformation of maize plants was carried out as in Pareddy, D., Petolino, J., Skokut, T., Hopkins, N., Miller, M., Welter, M., Smith, K., Clayton, D., Pescitelli, S ., Gould, A., Maize Transformation via helium blasting. Maydica. 42 (2): 143-154, 1997.
Beispiel 10Example 10
Erzeugung transgener Pflanzen, welche ein Enzym mit Dihydrooro- tase-Aktivität überexprimieren.Generation of transgenic plants which overexpress an enzyme with dihydroorotase activity.
Es wurde eine cDNA hergestellt, die für ein Enzym mit Dihydrooro- tase-Aktivität aus E.coli codiert, das an eine zum Import des Proteins in die Plastiden führende Signalsequenz (entnommen einem Enzym mit Tranketolase-Aktivität aus Tabak) fusioniert wurde. Hierzu wurde zunächst anhand der cDNA für die Dihydroorotase aus E.coli (Genbank Acc. Nr. X04469) die Oligonukleotide 5'-GTCGACAT- GACTGCACCATCCCAGG-3' und 5 ' -CGATTTTTATTGTTTAACGGACC-3 ' für eine Polymerase Kettenreakion (PCR) abgeleitet. Durch eine PCR mit genomischer DNA aus E.coli XL-1 blue als Matrize wurde eine Sall- Restriktionsschnittstelle vor dem Startcodon der Dihydroorotase cDNA eingeführt. Die Reaktionsgemische enthielten ca. 1 ng/μl Matrizen DNA, 0,5 μM der Oligonukleotide und, 200 μM Desoxy-Nukleo- tide (Pharmacia), 50 mM KCl, 10 mM Tris-HCl (pH 8,3 bei 25 °C, 1,5 mM MgCl ) und 0.02 U/μl Pwo Polymerase (Boehringer Mannheim) und wurden in einer PCR-Maschine der Firma Perkin Eimer mit folgendem Temperaturprogramm inkubiert:A cDNA was produced which codes for an enzyme with dihydroorotase activity from E. coli which was fused to a signal sequence leading to the import of the protein into the plastids (taken from an enzyme with tranketolase activity from tobacco). For this purpose, the oligonucleotides 5'-GTCGACAT-GACTGCACCATCCCAGG-3 'and 5' -CGATTTTTATTGTTTAACGGACC-3 'for a polymerase chain reaction (PCR) were first derived using the cDNA for the dihydroorotase from E. coli (Genbank Acc. No. X04469). A Sall- was identified by PCR with genomic DNA from E.coli XL-1 blue as a template. Restriction site introduced before the start codon of the dihydroorotase cDNA. The reaction mixtures contained approx. 1 ng / μl of template DNA, 0.5 μM of the oligonucleotides and, 200 μM deoxy nucleotides (Pharmacia), 50 mM KCl, 10 mM Tris-HCl (pH 8.3 at 25 ° C. 1.5 mM MgCl) and 0.02 U / μl Pwo polymerase (Boehringer Mannheim) and were incubated in a PCR machine from Perkin Elmer with the following temperature program:
Anlagerungstemperatur: 50°C, 45 sec Denaturierungstemperatur : 95°C, 45 secAnnealing temperature: 50 ° C, 45 sec. Denaturation temperature: 95 ° C, 45 sec
Elongationstemperatur: 72°C, 120 secElongation temperature: 72 ° C, 120 sec
Anzahl der Zyklen: 30Number of cycles: 30
Das erhaltene Fragment von 1059 bp wurde in den mit EcoRV gespal- tenen Vektor pBluescript SK- (Stratagene) ligiert. Durch Kon- trollspaltung wurde ein Klon identifiziert Kl, dessen Insert durch Sall in voller Länge exzisierbar ist (1059 bp + 18 bp der "muliple cloning site" des Vektors) .The 1059 bp fragment obtained was ligated into the vector pBluescript SK- (Stratagene), which had been split with EcoRV. A clone was identified by control cleavage, the insert of which can be excised in full length by Sall (1059 bp + 18 bp of the "multiple cloning site" of the vector).
Zur Überprüfung der Funktionalität des codierten Enzyms, wurde das 1077 bp Sall-Fragment aus Kl in den Expressionsvector pQE-9 (Quiagen) ligiert. Die korrekte Orientierung des Fragmentes wurde durch Restriktionsspaltung mit BamHI kontrolliert. Mit dem erhaltenen Konstrukt K2 wurde die pyrC E.coli Mutante CGSC#5152 (E.coli genetic stock center, York) transformiert. Die Transformanden wuchsen auf M9-Minimalmedien mit 20mg/l Methionin ohne Uridin, während Mutanten, die mit dem leeren pQE-9 Vektor transformiert wurden unter diesen Bedingungen kein Wachstum zeigten.To check the functionality of the encoded enzyme, the 1077 bp Sall fragment from Kl was ligated into the expression vector pQE-9 (Quiagen). The correct orientation of the fragment was checked by restriction cleavage with BamHI. The pyrC E. coli mutant CGSC # 5152 (E. coli genetic stock center, York) was transformed with the construct K2 obtained. The transformants grew on M9 minimal media with 20 mg / l methionine without uridine, while mutants transformed with the empty pQE-9 vector showed no growth under these conditions.
Für die Transformation von Pflanzen wurde ein Transfervektor erzeugt, indem das 1077 bp Sall-Fragment aus Kl in den mit Sall gespaltenen Vektor pTK-TP-BinAR9 (R. Badur, 1998 Doktorarbeit, Universität Göttingen) ligiert wurde. Auf diese Weise wurde eine Fusion des Leserasters der Dihydroorotase aus E.coli an ein pla- stidäres Transitpeptid, bestehend aus den N-terminalen 60 Aminosäuren der Transketolase aus Tabak (Genbank Acc. #CAA03393) erreicht (Konstrukt K3, Abb. 5) . Die fusionierte cDNA Sequenz steht unter Kontrolle des Blumenkohlmosaik-Virus 35S-Promoters und des Octopinsynthase-Terminators aus Agrobacterium tumefaciens.A transfer vector was generated for the transformation of plants by ligating the 1077 bp Sall fragment from Kl into the vector pTK-TP-BinAR9 cleaved with Sall (R. Badur, 1998 doctoral thesis, University of Göttingen). In this way a fusion of the reading frame of the dihydroorotase from E. coli to a plastid transit peptide, consisting of the N-terminal 60 amino acids of the transketolase from tobacco (Genbank Acc. # CAA03393) was achieved (construct K3, Fig. 5). The fused cDNA sequence is under the control of the cauliflower mosaic virus 35S promoter and the octopine synthase terminator from Agrobacterium tumefaciens.
Das Konstrukt K3 wurde zur Transformation von Tabak, Arabidopsis thaliana und Kartoffelpflanzen eingesetzt.The construct K3 was used to transform tobacco, Arabidopsis thaliana and potato plants.
Regenerierte Sprosse wurden auf 2MS-Medium mit Kanamycin und Cla- foran erhalten, nach Bewurzelung in Erde überführt und nach Kultivierung für zwei Wochen in einer Klimakammer oder im Gewächshaus (wie oben beschrieben) auf Dihydroorotase-Expression mittels Northern-blot Analyse untersucht. Linien mit erhöhten RNA-Spie- geln der Dihydroorotase wurden auf veränderte Metabolit- und Stärkegehalte in Blattgeweben bzw. Knollen untersucht. Es ließ sich in den transgenen Linien ein erhöhter Gehalt an Uridinnu- i kleotiden und ein erhöhter Stärkegehalt im Vergleich zu untrans - formierten Kontrollpflanzen feststellen.Regenerated shoots were obtained on 2MS medium with kanamycin and claforan, transferred to soil after rooting and after cultivation for two weeks in a climatic chamber or in the greenhouse (as described above) for dihydroorotase expression Northern blot analysis examined. Lines with increased RNA levels of dihydroorotase were examined for altered metabolite and starch contents in leaf tissues or tubers. An increased uridine nucleotide content and an increased starch content were found in the transgenic lines compared to untransformed control plants.
00
55
00
55
00
5 5
Claims
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| AU76470/00A AU7647000A (en) | 1999-08-20 | 2000-08-12 | Increasing the polysaccharide content in plants |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| DE19939688 | 1999-08-20 | ||
| DE19939688.4 | 1999-08-20 |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| WO2001014569A2 true WO2001014569A2 (en) | 2001-03-01 |
| WO2001014569A3 WO2001014569A3 (en) | 2001-10-11 |
Family
ID=7919149
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/EP2000/007884 Ceased WO2001014569A2 (en) | 1999-08-20 | 2000-08-12 | Increasing the polysaccharide content in plants |
Country Status (3)
| Country | Link |
|---|---|
| AR (1) | AR026151A1 (en) |
| AU (1) | AU7647000A (en) |
| WO (1) | WO2001014569A2 (en) |
Cited By (176)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2001018190A3 (en) * | 1999-09-07 | 2001-10-11 | Basf Ag | Dihydroorotase extracted from plants |
| WO2001059089A3 (en) * | 2000-02-08 | 2002-07-04 | Genentech Inc | Improved galactosylation of recombinant glycoproteins |
| EP2039771A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| EP2039772A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants introduction |
| EP2039770A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| EP2072506A1 (en) | 2007-12-21 | 2009-06-24 | Bayer CropScience AG | Thiazolyloxyphenylamidine or thiadiazolyloxyphenylamidine und its use as fungicide |
| EP2090168A1 (en) | 2008-02-12 | 2009-08-19 | Bayer CropScience AG | Method for improving plant growth |
| EP2168434A1 (en) | 2008-08-02 | 2010-03-31 | Bayer CropScience AG | Use of azols to increase resistance of plants of parts of plants to abiotic stress |
| EP2198709A1 (en) | 2008-12-19 | 2010-06-23 | Bayer CropScience AG | Method for treating resistant animal pests |
| EP2201838A1 (en) | 2008-12-05 | 2010-06-30 | Bayer CropScience AG | Active ingredient-beneficial organism combinations with insecticide and acaricide properties |
| EP2204094A1 (en) | 2008-12-29 | 2010-07-07 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants Introduction |
| WO2010083955A2 (en) | 2009-01-23 | 2010-07-29 | Bayer Cropscience Aktiengesellschaft | Use of enaminocarboxylic compounds for fighting viruses transmitted by insects |
| WO2010086095A1 (en) | 2009-01-29 | 2010-08-05 | Bayer Cropscience Ag | Method for improved utilization of the production potential of transgenic plants introduction |
| WO2010086311A1 (en) | 2009-01-28 | 2010-08-05 | Bayer Cropscience Ag | Fungicide n-cycloalkyl-n-bicyclicmethylene-carboxamide derivatives |
| EP2218717A1 (en) | 2009-02-17 | 2010-08-18 | Bayer CropScience AG | Fungicidal N-((HET)Arylethyl)thiocarboxamide derivatives |
| WO2010094728A1 (en) | 2009-02-19 | 2010-08-26 | Bayer Cropscience Ag | Pesticide composition comprising a tetrazolyloxime derivative and a fungicide or an insecticide active substance |
| WO2010094666A2 (en) | 2009-02-17 | 2010-08-26 | Bayer Cropscience Ag | Fungicidal n-(phenylcycloalkyl)carboxamide, n-(benzylcycloalkyl)carboxamide and thiocarboxamide derivatives |
| EP2223602A1 (en) | 2009-02-23 | 2010-09-01 | Bayer CropScience AG | Method for improved utilisation of the production potential of genetically modified plants |
| EP2232995A1 (en) | 2009-03-25 | 2010-09-29 | Bayer CropScience AG | Method for improved utilisation of the production potential of transgenic plants |
| EP2239331A1 (en) | 2009-04-07 | 2010-10-13 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| EP2251331A1 (en) | 2009-05-15 | 2010-11-17 | Bayer CropScience AG | Fungicide pyrazole carboxamides derivatives |
| EP2255626A1 (en) | 2009-05-27 | 2010-12-01 | Bayer CropScience AG | Use of succinate dehydrogenase inhibitors to increase resistance of plants or parts of plants to abiotic stress |
| WO2011015524A2 (en) | 2009-08-03 | 2011-02-10 | Bayer Cropscience Ag | Fungicide heterocycles derivatives |
| EP2292094A1 (en) | 2009-09-02 | 2011-03-09 | Bayer CropScience AG | Active compound combinations |
| WO2011080256A1 (en) | 2009-12-28 | 2011-07-07 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2011080254A2 (en) | 2009-12-28 | 2011-07-07 | Bayer Cropscience Ag | Fungicide hydroximoyl-heterocycles derivatives |
| WO2011080255A2 (en) | 2009-12-28 | 2011-07-07 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| EP2343280A1 (en) | 2009-12-10 | 2011-07-13 | Bayer CropScience AG | Fungicide quinoline derivatives |
| WO2011089071A2 (en) | 2010-01-22 | 2011-07-28 | Bayer Cropscience Ag | Acaricide and/or insecticide active substance combinations |
| WO2011107504A1 (en) | 2010-03-04 | 2011-09-09 | Bayer Cropscience Ag | Fluoroalkyl-substituted 2-amidobenzimidazoles and the use thereof for boosting stress tolerance in plants |
| EP2374791A1 (en) | 2008-08-14 | 2011-10-12 | Bayer CropScience Aktiengesellschaft | Insecticidal 4-phenyl-1H pyrazoles |
| WO2011124553A2 (en) | 2010-04-09 | 2011-10-13 | Bayer Cropscience Ag | Use of derivatives of the (1-cyanocyclopropyl)phenylphosphinic acid, the esters thereof and/or the salts thereof for enhancing the tolerance of plants to abiotic stress |
| WO2011124554A2 (en) | 2010-04-06 | 2011-10-13 | Bayer Cropscience Ag | Use of 4-phenylbutyric acid and/or the salts thereof for enhancing the stress tolerance of plants |
| WO2011134911A2 (en) | 2010-04-28 | 2011-11-03 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2011134913A1 (en) | 2010-04-28 | 2011-11-03 | Bayer Cropscience Ag | Fungicide hydroximoyl-heterocycles derivatives |
| WO2011134912A1 (en) | 2010-04-28 | 2011-11-03 | Bayer Cropscience Ag | Fungicide hydroximoyl-heterocycles derivatives |
| WO2011151369A1 (en) | 2010-06-03 | 2011-12-08 | Bayer Cropscience Ag | N-[(het)arylethyl)] pyrazole(thio)carboxamides and their heterosubstituted analogues |
| WO2011151368A2 (en) | 2010-06-03 | 2011-12-08 | Bayer Cropscience Ag | Fungicide n-[(trisubstitutedsilyl)methyl]-carboxamide derivatives |
| WO2011151370A1 (en) | 2010-06-03 | 2011-12-08 | Bayer Cropscience Ag | N-[(het)arylalkyl)] pyrazole (thio)carboxamides and their heterosubstituted analogues |
| WO2011154159A1 (en) | 2010-06-09 | 2011-12-15 | Bayer Bioscience N.V. | Methods and means to modify a plant genome at a nucleotide sequence commonly used in plant genome engineering |
| WO2011154158A1 (en) | 2010-06-09 | 2011-12-15 | Bayer Bioscience N.V. | Methods and means to modify a plant genome at a nucleotide sequence commonly used in plant genome engineering |
| US8080688B2 (en) | 2007-03-12 | 2011-12-20 | Bayer Cropscience Ag | 3, 4-disubstituted phenoxyphenylamidines and use thereof as fungicides |
| WO2012010579A2 (en) | 2010-07-20 | 2012-01-26 | Bayer Cropscience Ag | Benzocycloalkenes as antifungal agents |
| WO2012028578A1 (en) | 2010-09-03 | 2012-03-08 | Bayer Cropscience Ag | Substituted fused pyrimidinones and dihydropyrimidinones |
| WO2012038476A1 (en) | 2010-09-22 | 2012-03-29 | Bayer Cropscience Ag | Use of active ingredients for controlling nematodes in nematode-resistant crops |
| WO2012045798A1 (en) | 2010-10-07 | 2012-04-12 | Bayer Cropscience Ag | Fungicide composition comprising a tetrazolyloxime derivative and a thiazolylpiperidine derivative |
| WO2012052490A1 (en) | 2010-10-21 | 2012-04-26 | Bayer Cropscience Ag | N-benzyl heterocyclic carboxamides |
| WO2012052489A1 (en) | 2010-10-21 | 2012-04-26 | Bayer Cropscience Ag | 1-(heterocyclic carbonyl) piperidines |
| US8168567B2 (en) | 2007-04-19 | 2012-05-01 | Bayer Cropscience Ag | Thiadiazolyl oxyphenyl amidines and the use thereof as a fungicide |
| WO2012059497A1 (en) | 2010-11-02 | 2012-05-10 | Bayer Cropscience Ag | N-hetarylmethyl pyrazolylcarboxamides |
| WO2012065945A1 (en) | 2010-11-15 | 2012-05-24 | Bayer Cropscience Ag | 5-halogenopyrazole(thio)carboxamides |
| WO2012065944A1 (en) | 2010-11-15 | 2012-05-24 | Bayer Cropscience Ag | N-aryl pyrazole(thio)carboxamides |
| WO2012065947A1 (en) | 2010-11-15 | 2012-05-24 | Bayer Cropscience Ag | 5-halogenopyrazolecarboxamides |
| EP2460407A1 (en) | 2010-12-01 | 2012-06-06 | Bayer CropScience AG | Agent combinations comprising pyridylethyl benzamides and other agents |
| EP2460406A1 (en) | 2010-12-01 | 2012-06-06 | Bayer CropScience AG | Use of fluopyram for controlling nematodes in nematode resistant crops |
| WO2012072660A1 (en) | 2010-12-01 | 2012-06-07 | Bayer Cropscience Ag | Use of fluopyram for controlling nematodes in crops and for increasing yield |
| WO2012089757A1 (en) | 2010-12-29 | 2012-07-05 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2012089721A1 (en) | 2010-12-30 | 2012-07-05 | Bayer Cropscience Ag | Use of substituted spirocyclic sulfonamidocarboxylic acids, carboxylic esters thereof, carboxamides thereof and carbonitriles thereof or salts thereof for enhancement of stress tolerance in plants |
| EP2474542A1 (en) | 2010-12-29 | 2012-07-11 | Bayer CropScience AG | Fungicide hydroximoyl-tetrazole derivatives |
| EP2494867A1 (en) | 2011-03-01 | 2012-09-05 | Bayer CropScience AG | Halogen-substituted compounds in combination with fungicides |
| WO2012120105A1 (en) | 2011-03-10 | 2012-09-13 | Bayer Cropscience Ag | Use of lipochito-oligosaccharide compounds for safeguarding seed safety of treated seeds |
| WO2012123434A1 (en) | 2011-03-14 | 2012-09-20 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2012136581A1 (en) | 2011-04-08 | 2012-10-11 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| US8288426B2 (en) | 2006-12-22 | 2012-10-16 | Bayer Cropscience Ag | Pesticidal composition comprising fenamidone and an insecticide compound |
| EP2511255A1 (en) | 2011-04-15 | 2012-10-17 | Bayer CropScience AG | Substituted prop-2-in-1-ol and prop-2-en-1-ol derivatives |
| WO2012139891A1 (en) | 2011-04-15 | 2012-10-18 | Bayer Cropscience Ag | Substituted vinyl and alkinyl cyclohexenols as active agents against abiotic stress in plants |
| WO2012139890A1 (en) | 2011-04-15 | 2012-10-18 | Bayer Cropscience Ag | Substituted 5-(cyclohex-2-en-1-yl)-penta-2,4-dienes and 5-(cyclohex-2-en-1-yl)-pent-2-en-4-ines as active agents against abiotic stress in plants |
| WO2012139892A1 (en) | 2011-04-15 | 2012-10-18 | Bayer Cropscience Ag | Substituted 5-(bicyclo[4.1.0]hept-3-en-2-yl)-penta-2,4-dienes and 5-(bicyclo[4.1.0]hept-3-en-2-yl)-pent-2-ene-4-ines as active agents against abiotic stress in plants |
| US8299302B2 (en) | 2007-03-12 | 2012-10-30 | Bayer Cropscience Ag | 4-Cycloalkyl or 4-substituted phenoxyphenylamidines and use thereof as fungicides |
| WO2012168124A1 (en) | 2011-06-06 | 2012-12-13 | Bayer Cropscience Nv | Methods and means to modify a plant genome at a preselected site |
| US8334237B2 (en) | 2007-03-12 | 2012-12-18 | Bayer Cropscience Ag | Substituted phenylamidines and the use thereof as fungicides |
| WO2013004652A1 (en) | 2011-07-04 | 2013-01-10 | Bayer Intellectual Property Gmbh | Use of substituted isoquinolinones, isoquinolindiones, isoquinolintriones and dihydroisoquinolinones or in each case salts thereof as active agents against abiotic stress in plants |
| WO2013020985A1 (en) | 2011-08-10 | 2013-02-14 | Bayer Intellectual Property Gmbh | Active compound combinations comprising specific tetramic acid derivatives |
| EP2561759A1 (en) | 2011-08-26 | 2013-02-27 | Bayer Cropscience AG | Fluoroalkyl-substituted 2-amidobenzimidazoles and their effect on plant growth |
| WO2013026740A2 (en) | 2011-08-22 | 2013-02-28 | Bayer Cropscience Nv | Methods and means to modify a plant genome |
| WO2013026836A1 (en) | 2011-08-22 | 2013-02-28 | Bayer Intellectual Property Gmbh | Fungicide hydroximoyl-tetrazole derivatives |
| US8394991B2 (en) | 2007-03-12 | 2013-03-12 | Bayer Cropscience Ag | Phenoxy substituted phenylamidine derivatives and their use as fungicides |
| WO2013034621A1 (en) | 2011-09-09 | 2013-03-14 | Bayer Intellectual Property Gmbh | Acyl-homoserine lactone derivatives for improving plant yield |
| WO2013037958A1 (en) | 2011-09-16 | 2013-03-21 | Bayer Intellectual Property Gmbh | Use of phenylpyrazolin-3-carboxylates for improving plant yield |
| WO2013037717A1 (en) | 2011-09-12 | 2013-03-21 | Bayer Intellectual Property Gmbh | Fungicidal 4-substituted-3-{phenyl[(heterocyclylmethoxy)imino]methyl}-1,2,4-oxadizol-5(4h)-one derivatives |
| WO2013037955A1 (en) | 2011-09-16 | 2013-03-21 | Bayer Intellectual Property Gmbh | Use of acylsulfonamides for improving plant yield |
| WO2013037956A1 (en) | 2011-09-16 | 2013-03-21 | Bayer Intellectual Property Gmbh | Use of 5-phenyl- or 5-benzyl-2 isoxazoline-3 carboxylates for improving plant yield |
| WO2013041602A1 (en) | 2011-09-23 | 2013-03-28 | Bayer Intellectual Property Gmbh | Use of 4-substituted 1-phenyl-pyrazole-3-carboxylic-acid derivatives as agents against abiotic plant stress |
| WO2013050410A1 (en) | 2011-10-04 | 2013-04-11 | Bayer Intellectual Property Gmbh | RNAi FOR THE CONTROL OF FUNGI AND OOMYCETES BY INHIBITING SACCHAROPINE DEHYDROGENASE GENE |
| WO2013050324A1 (en) | 2011-10-06 | 2013-04-11 | Bayer Intellectual Property Gmbh | Combination, containing 4-phenylbutyric acid (4-pba) or a salt thereof (component (a)) and one or more selected additional agronomically active compounds (component(s) (b)), that reduces abiotic plant stress |
| WO2013075817A1 (en) | 2011-11-21 | 2013-05-30 | Bayer Intellectual Property Gmbh | Fungicide n-[(trisubstitutedsilyl)methyl]-carboxamide derivatives |
| US8455480B2 (en) | 2007-09-26 | 2013-06-04 | Bayer Cropscience Ag | Active agent combinations having insecticidal and acaricidal properties |
| WO2013079566A2 (en) | 2011-11-30 | 2013-06-06 | Bayer Intellectual Property Gmbh | Fungicidal n-bicycloalkyl and n-tricycloalkyl (thio)carboxamide derivatives |
| WO2013092519A1 (en) | 2011-12-19 | 2013-06-27 | Bayer Cropscience Ag | Use of anthranilic acid diamide derivatives for pest control in transgenic crops |
| WO2013098146A1 (en) | 2011-12-29 | 2013-07-04 | Bayer Intellectual Property Gmbh | Fungicidal 3-[(1,3-thiazol-4-ylmethoxyimino)(phenyl)methyl]-2-substituted-1,2,4-oxadiazol-5(2h)-one derivatives |
| WO2013098147A1 (en) | 2011-12-29 | 2013-07-04 | Bayer Intellectual Property Gmbh | Fungicidal 3-[(pyridin-2-ylmethoxyimino)(phenyl)methyl]-2-substituted-1,2,4-oxadiazol-5(2h)-one derivatives |
| US8487118B2 (en) | 2009-01-19 | 2013-07-16 | Bayer Cropscience Ag | Cyclic diones and their use as insecticides, acaricides and/or fungicides |
| WO2013124275A1 (en) | 2012-02-22 | 2013-08-29 | Bayer Cropscience Ag | Use of succinate dehydrogenase inhibitors (sdhis) for controlling wood diseases in grape. |
| WO2013127704A1 (en) | 2012-02-27 | 2013-09-06 | Bayer Intellectual Property Gmbh | Active compound combinations containing a thiazoylisoxazoline and a fungicide |
| WO2013139949A1 (en) | 2012-03-23 | 2013-09-26 | Bayer Intellectual Property Gmbh | Compositions comprising a strigolactame compound for enhanced plant growth and yield |
| WO2013153143A1 (en) | 2012-04-12 | 2013-10-17 | Bayer Cropscience Ag | N-acyl- 2 - (cyclo) alkylpyrrolidines and piperidines useful as fungicides |
| WO2013156559A1 (en) | 2012-04-20 | 2013-10-24 | Bayer Cropscience Ag | N-cycloalkyl-n-[(heterocyclylphenyl)methylene]-(thio)carboxamide derivatives |
| WO2013156560A1 (en) | 2012-04-20 | 2013-10-24 | Bayer Cropscience Ag | N-cycloalkyl-n-[(trisubstitutedsilylphenyl)methylene]-(thio)carboxamide derivatives |
| WO2013160230A1 (en) | 2012-04-23 | 2013-10-31 | Bayer Cropscience Nv | Targeted genome engineering in plants |
| EP2662361A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazol indanyl carboxamides |
| EP2662364A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazole tetrahydronaphthyl carboxamides |
| EP2662370A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole benzofuranyl carboxamides |
| EP2662363A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole biphenylcarboxamides |
| EP2662362A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazole indanyl carboxamides |
| EP2662360A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole indanyl carboxamides |
| WO2013167545A1 (en) | 2012-05-09 | 2013-11-14 | Bayer Cropscience Ag | Pyrazole indanyl carboxamides |
| WO2013167544A1 (en) | 2012-05-09 | 2013-11-14 | Bayer Cropscience Ag | 5-halogenopyrazole indanyl carboxamides |
| WO2013174836A1 (en) | 2012-05-22 | 2013-11-28 | Bayer Cropscience Ag | Active compounds combinations comprising a lipo-chitooligosaccharide derivative and a nematicide, insecticidal or fungicidal compound |
| WO2014009322A1 (en) | 2012-07-11 | 2014-01-16 | Bayer Cropscience Ag | Use of fungicidal combinations for increasing the tolerance of a plant towards abiotic stress |
| WO2014037340A1 (en) | 2012-09-05 | 2014-03-13 | Bayer Cropscience Ag | Use of substituted 2-amidobenzimidazoles, 2-amidobenzoxazoles and 2-amidobenzothiazoles or salts thereof as active substances against abiotic plant stress |
| WO2014060518A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Method of plant growth promotion using carboxamide derivatives |
| WO2014060519A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Method for enhancing tolerance to abiotic stress in plants using carboxamide or thiocarboxamide derivatives |
| WO2014060520A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Method for treating plants against fungi resistant to fungicides using carboxamide or thiocarboxamide derivatives |
| WO2014060502A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Active compound combinations comprising carboxamide derivatives |
| EP2735231A1 (en) | 2012-11-23 | 2014-05-28 | Bayer CropScience AG | Active compound combinations |
| WO2014079957A1 (en) | 2012-11-23 | 2014-05-30 | Bayer Cropscience Ag | Selective inhibition of ethylene signal transduction |
| WO2014083033A1 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropsience Ag | Binary fungicidal or pesticidal mixture |
| WO2014083089A1 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Ternary fungicidal and pesticidal mixtures |
| WO2014083031A2 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Binary pesticidal and fungicidal mixtures |
| WO2014082950A1 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Ternary fungicidal mixtures |
| WO2014083088A2 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Binary fungicidal mixtures |
| EP2740720A1 (en) | 2012-12-05 | 2014-06-11 | Bayer CropScience AG | Substituted bicyclic and tricyclic pent-2-en-4-inic acid derivatives and their use for enhancing the stress tolerance in plants |
| EP2740356A1 (en) | 2012-12-05 | 2014-06-11 | Bayer CropScience AG | Substituted (2Z)-5(1-Hydroxycyclohexyl)pent-2-en-4-inic acid derivatives |
| WO2014086751A1 (en) | 2012-12-05 | 2014-06-12 | Bayer Cropscience Ag | Use of substituted 1-(aryl ethynyl)-, 1-(heteroaryl ethynyl)-, 1-(heterocyclyl ethynyl)- and 1-(cyloalkenyl ethynyl)-cyclohexanols as active agents against abiotic plant stress |
| WO2014090765A1 (en) | 2012-12-12 | 2014-06-19 | Bayer Cropscience Ag | Use of 1-[2-fluoro-4-methyl-5-(2,2,2-trifluoroethylsulfinyl)phenyl]-5-amino-3-trifluoromethyl)-1 h-1,2,4 tfia zole for controlling nematodes in nematode-resistant crops |
| WO2014095677A1 (en) | 2012-12-19 | 2014-06-26 | Bayer Cropscience Ag | Difluoromethyl-nicotinic- tetrahydronaphtyl carboxamides |
| WO2014095826A1 (en) | 2012-12-18 | 2014-06-26 | Bayer Cropscience Ag | Binary fungicidal and bactericidal combinations |
| US8796175B2 (en) | 2008-08-29 | 2014-08-05 | Bayer Cropscience Ag | Method for enhancing plant intrinsic defense |
| US8828907B2 (en) | 2009-03-25 | 2014-09-09 | Bayer Cropscience Ag | Active ingredient combinations having insecticidal and acaricidal properties |
| US8828906B2 (en) | 2009-03-25 | 2014-09-09 | Bayer Cropscience Ag | Active compound combinations having insecticidal and acaricidal properties |
| WO2014135608A1 (en) | 2013-03-07 | 2014-09-12 | Bayer Cropscience Ag | Fungicidal 3-{phenyl[(heterocyclylmethoxy)imino]methyl}-heterocycle derivatives |
| US8835657B2 (en) | 2009-05-06 | 2014-09-16 | Bayer Cropscience Ag | Cyclopentanedione compounds and their use as insecticides, acaricides and/or fungicides |
| US8846568B2 (en) | 2009-03-25 | 2014-09-30 | Bayer Cropscience Ag | Active compound combinations having insecticidal and acaricidal properties |
| US8846567B2 (en) | 2009-03-25 | 2014-09-30 | Bayer Cropscience Ag | Active compound combinations having insecticidal and acaricidal properties |
| WO2014161821A1 (en) | 2013-04-02 | 2014-10-09 | Bayer Cropscience Nv | Targeted genome engineering in eukaryotes |
| WO2014167008A1 (en) | 2013-04-12 | 2014-10-16 | Bayer Cropscience Ag | Novel triazolinthione derivatives |
| WO2014167009A1 (en) | 2013-04-12 | 2014-10-16 | Bayer Cropscience Ag | Novel triazole derivatives |
| WO2014170345A2 (en) | 2013-04-19 | 2014-10-23 | Bayer Cropscience Ag | Method for improved utilization of the production potential of transgenic plants |
| WO2014170364A1 (en) | 2013-04-19 | 2014-10-23 | Bayer Cropscience Ag | Binary insecticidal or pesticidal mixture |
| WO2014177514A1 (en) | 2013-04-30 | 2014-11-06 | Bayer Cropscience Ag | Nematicidal n-substituted phenethylcarboxamides |
| WO2014177582A1 (en) | 2013-04-30 | 2014-11-06 | Bayer Cropscience Ag | N-(2-fluoro-2-phenethyl)carboxamides as nematicides and endoparasiticides |
| WO2014206953A1 (en) | 2013-06-26 | 2014-12-31 | Bayer Cropscience Ag | N-cycloalkyl-n-[(bicyclylphenyl)methylene]-(thio)carboxamide derivatives |
| US8927583B2 (en) | 2006-12-22 | 2015-01-06 | Bayer Cropscience Ag | Pesticidal composition comprising a 2-pyrdilmethylbenzamide derivative and an insecticide compound |
| WO2015004040A1 (en) | 2013-07-09 | 2015-01-15 | Bayer Cropscience Ag | Use of selected pyridone carboxamides or salts thereof as active substances against abiotic plant stress |
| US9012360B2 (en) | 2009-03-25 | 2015-04-21 | Bayer Intellectual Property Gmbh | Synergistic combinations of active ingredients |
| WO2015082586A1 (en) | 2013-12-05 | 2015-06-11 | Bayer Cropscience Ag | N-cycloalkyl-n-{[2-(1-substitutedcycloalkyl)phenyl]methylene}-(thio)carboxamide derivatives |
| WO2015082587A1 (en) | 2013-12-05 | 2015-06-11 | Bayer Cropscience Ag | N-cycloalkyl-n-{[2-(1-substitutedcycloalkyl)phenyl]methylene}-(thio)carboxamide derivatives |
| US9199922B2 (en) | 2007-03-12 | 2015-12-01 | Bayer Intellectual Property Gmbh | Dihalophenoxyphenylamidines and use thereof as fungicides |
| US9232794B2 (en) | 2009-06-02 | 2016-01-12 | Bayer Intellectual Property Gmbh | Use of succinate dehydrogenase inhibitors for controlling Sclerotinia ssp |
| WO2016012362A1 (en) | 2014-07-22 | 2016-01-28 | Bayer Cropscience Aktiengesellschaft | Substituted cyano cycloalkyl penta-2,4-dienes, cyano cycloalkyl pent-2-en-4-ynes, cyano heterocyclyl penta-2,4-dienes and cyano heterocyclyl pent-2-en-4-ynes as active substances against abiotic plant stress |
| EP2997825A1 (en) | 2011-04-22 | 2016-03-23 | Bayer Intellectual Property GmbH | Active compound combinations comprising a (thio)carboxamide derivative and a fungicidal compound |
| EP3000809A1 (en) | 2009-05-15 | 2016-03-30 | Bayer Intellectual Property GmbH | Fungicide pyrazole carboxamides derivatives |
| WO2016096942A1 (en) | 2014-12-18 | 2016-06-23 | Bayer Cropscience Aktiengesellschaft | Use of selected pyridone carboxamides or salts thereof as active substances against abiotic plant stress |
| WO2016166077A1 (en) | 2015-04-13 | 2016-10-20 | Bayer Cropscience Aktiengesellschaft | N-cycloalkyl-n-(biheterocyclyethylene)-(thio)carboxamide derivatives |
| US9763451B2 (en) | 2008-12-29 | 2017-09-19 | Bayer Intellectual Property Gmbh | Method for improved use of the production potential of genetically modified plants |
| WO2018019676A1 (en) | 2016-07-29 | 2018-02-01 | Bayer Cropscience Aktiengesellschaft | Active compound combinations and methods to protect the propagation material of plants |
| WO2018054832A1 (en) | 2016-09-22 | 2018-03-29 | Bayer Cropscience Aktiengesellschaft | Novel triazole derivatives |
| WO2018054911A1 (en) | 2016-09-23 | 2018-03-29 | Bayer Cropscience Nv | Targeted genome optimization in plants |
| WO2018054829A1 (en) | 2016-09-22 | 2018-03-29 | Bayer Cropscience Aktiengesellschaft | Novel triazole derivatives and their use as fungicides |
| WO2018077711A2 (en) | 2016-10-26 | 2018-05-03 | Bayer Cropscience Aktiengesellschaft | Use of pyraziflumid for controlling sclerotinia spp in seed treatment applications |
| EP3332645A1 (en) | 2016-12-12 | 2018-06-13 | Bayer Cropscience AG | Use of substituted pyrimidine diones or their salts as agents to combat abiotic plant stress |
| WO2018104392A1 (en) | 2016-12-08 | 2018-06-14 | Bayer Cropscience Aktiengesellschaft | Use of insecticides for controlling wireworms |
| WO2018108627A1 (en) | 2016-12-12 | 2018-06-21 | Bayer Cropscience Aktiengesellschaft | Use of substituted indolinylmethyl sulfonamides, or the salts thereof for increasing the stress tolerance of plants |
| DE102007045953B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Drug combinations with insecticidal and acaricidal properties |
| DE102007045919B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Drug combinations with insecticidal and acaricidal properties |
| DE102007045920B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Synergistic drug combinations |
| WO2019025153A1 (en) | 2017-07-31 | 2019-02-07 | Bayer Cropscience Aktiengesellschaft | USE OF SUBSTITUTED N-SULFONYL-N'-ARYLDIAMINOALKANES AND N-SULFONYL-N'-HETEROARYL DIAMINOALKANES OR THEIR SALTS TO INCREASE STRESSTOLERANCE IN PLANTS |
| WO2019060746A1 (en) | 2017-09-21 | 2019-03-28 | The Broad Institute, Inc. | Systems, methods, and compositions for targeted nucleic acid editing |
| WO2019126709A1 (en) | 2017-12-22 | 2019-06-27 | The Broad Institute, Inc. | Cas12b systems, methods, and compositions for targeted dna base editing |
| WO2019233863A1 (en) | 2018-06-04 | 2019-12-12 | Bayer Aktiengesellschaft | Herbicidally active bicyclic benzoylpyrazoles |
| WO2020131862A1 (en) | 2018-12-17 | 2020-06-25 | The Broad Institute, Inc. | Crispr-associated transposase systems and methods of use thereof |
| US10968257B2 (en) | 2018-04-03 | 2021-04-06 | The Broad Institute, Inc. | Target recognition motifs and uses thereof |
| US11180751B2 (en) | 2015-06-18 | 2021-11-23 | The Broad Institute, Inc. | CRISPR enzymes and systems |
| US11591601B2 (en) | 2017-05-05 | 2023-02-28 | The Broad Institute, Inc. | Methods for identification and modification of lncRNA associated with target genotypes and phenotypes |
| US12297436B2 (en) | 2017-05-18 | 2025-05-13 | The Broad Institute, Inc. | Systems, methods, and compositions for targeted nucleic acid editing |
| US12499971B2 (en) | 2016-09-28 | 2025-12-16 | The Broad Institute, Inc. | Systematic screening and mapping of regulatory elements in non-coding genomic regions, methods, compositions, and applications thereof |
Families Citing this family (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| DE102007045922A1 (en) | 2007-09-26 | 2009-04-02 | Bayer Cropscience Ag | Drug combinations with insecticidal and acaricidal properties |
Family Cites Families (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| ATE212670T1 (en) * | 1990-06-18 | 2002-02-15 | Monsanto Technology Llc | INCREASED STARCH CONTENT IN PLANTS |
| GB9307408D0 (en) * | 1993-04-08 | 1993-06-02 | Danisco | Transgenic plants |
| DE19501906A1 (en) * | 1995-01-23 | 1996-07-25 | Basf Ag | Transketolase |
-
2000
- 2000-08-12 AU AU76470/00A patent/AU7647000A/en not_active Abandoned
- 2000-08-12 WO PCT/EP2000/007884 patent/WO2001014569A2/en not_active Ceased
- 2000-08-22 AR ARP000104330 patent/AR026151A1/en unknown
Cited By (200)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US7320877B1 (en) | 1999-09-07 | 2008-01-22 | Basf Aktiengesellschaft | Dihydroorotase extracted from plants |
| US7727753B2 (en) | 1999-09-07 | 2010-06-01 | Basf Aktiengesellschaft | Plant dihydroorotase |
| WO2001018190A3 (en) * | 1999-09-07 | 2001-10-11 | Basf Ag | Dihydroorotase extracted from plants |
| WO2001059089A3 (en) * | 2000-02-08 | 2002-07-04 | Genentech Inc | Improved galactosylation of recombinant glycoproteins |
| US8288426B2 (en) | 2006-12-22 | 2012-10-16 | Bayer Cropscience Ag | Pesticidal composition comprising fenamidone and an insecticide compound |
| US8927583B2 (en) | 2006-12-22 | 2015-01-06 | Bayer Cropscience Ag | Pesticidal composition comprising a 2-pyrdilmethylbenzamide derivative and an insecticide compound |
| US9199922B2 (en) | 2007-03-12 | 2015-12-01 | Bayer Intellectual Property Gmbh | Dihalophenoxyphenylamidines and use thereof as fungicides |
| US8080688B2 (en) | 2007-03-12 | 2011-12-20 | Bayer Cropscience Ag | 3, 4-disubstituted phenoxyphenylamidines and use thereof as fungicides |
| US8394991B2 (en) | 2007-03-12 | 2013-03-12 | Bayer Cropscience Ag | Phenoxy substituted phenylamidine derivatives and their use as fungicides |
| US8334237B2 (en) | 2007-03-12 | 2012-12-18 | Bayer Cropscience Ag | Substituted phenylamidines and the use thereof as fungicides |
| US8299302B2 (en) | 2007-03-12 | 2012-10-30 | Bayer Cropscience Ag | 4-Cycloalkyl or 4-substituted phenoxyphenylamidines and use thereof as fungicides |
| US8748662B2 (en) | 2007-03-12 | 2014-06-10 | Bayer Cropscience Ag | 4-cycloalkyl or 4-aryl substituted phenoxyphenylamidines and use thereof as fungicides |
| US8785692B2 (en) | 2007-03-12 | 2014-07-22 | Bayer Cropscience Ag | Substituted phenylamidines and the use thereof as fungicides |
| US8168567B2 (en) | 2007-04-19 | 2012-05-01 | Bayer Cropscience Ag | Thiadiazolyl oxyphenyl amidines and the use thereof as a fungicide |
| DE102007045920B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Synergistic drug combinations |
| DE102007045919B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Drug combinations with insecticidal and acaricidal properties |
| US8455480B2 (en) | 2007-09-26 | 2013-06-04 | Bayer Cropscience Ag | Active agent combinations having insecticidal and acaricidal properties |
| DE102007045953B4 (en) | 2007-09-26 | 2018-07-05 | Bayer Intellectual Property Gmbh | Drug combinations with insecticidal and acaricidal properties |
| EP2072506A1 (en) | 2007-12-21 | 2009-06-24 | Bayer CropScience AG | Thiazolyloxyphenylamidine or thiadiazolyloxyphenylamidine und its use as fungicide |
| EP2090168A1 (en) | 2008-02-12 | 2009-08-19 | Bayer CropScience AG | Method for improving plant growth |
| EP2168434A1 (en) | 2008-08-02 | 2010-03-31 | Bayer CropScience AG | Use of azols to increase resistance of plants of parts of plants to abiotic stress |
| EP2374791A1 (en) | 2008-08-14 | 2011-10-12 | Bayer CropScience Aktiengesellschaft | Insecticidal 4-phenyl-1H pyrazoles |
| US8796175B2 (en) | 2008-08-29 | 2014-08-05 | Bayer Cropscience Ag | Method for enhancing plant intrinsic defense |
| EP2201838A1 (en) | 2008-12-05 | 2010-06-30 | Bayer CropScience AG | Active ingredient-beneficial organism combinations with insecticide and acaricide properties |
| EP2198709A1 (en) | 2008-12-19 | 2010-06-23 | Bayer CropScience AG | Method for treating resistant animal pests |
| EP2204094A1 (en) | 2008-12-29 | 2010-07-07 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants Introduction |
| US9763451B2 (en) | 2008-12-29 | 2017-09-19 | Bayer Intellectual Property Gmbh | Method for improved use of the production potential of genetically modified plants |
| WO2010075994A1 (en) | 2008-12-29 | 2010-07-08 | Bayer Cropscience Aktiengesellschaft | Treatment of transgenic crops with mixtures of fiproles and chloronicotinyls |
| EP2039772A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants introduction |
| EP2039771A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| EP2039770A2 (en) | 2009-01-06 | 2009-03-25 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| US8487118B2 (en) | 2009-01-19 | 2013-07-16 | Bayer Cropscience Ag | Cyclic diones and their use as insecticides, acaricides and/or fungicides |
| EP2227951A1 (en) | 2009-01-23 | 2010-09-15 | Bayer CropScience AG | Application of enaminocarbonyl compounds for combating viruses transmitted by insects |
| WO2010083955A2 (en) | 2009-01-23 | 2010-07-29 | Bayer Cropscience Aktiengesellschaft | Use of enaminocarboxylic compounds for fighting viruses transmitted by insects |
| WO2010086311A1 (en) | 2009-01-28 | 2010-08-05 | Bayer Cropscience Ag | Fungicide n-cycloalkyl-n-bicyclicmethylene-carboxamide derivatives |
| WO2010086095A1 (en) | 2009-01-29 | 2010-08-05 | Bayer Cropscience Ag | Method for improved utilization of the production potential of transgenic plants introduction |
| WO2010094666A2 (en) | 2009-02-17 | 2010-08-26 | Bayer Cropscience Ag | Fungicidal n-(phenylcycloalkyl)carboxamide, n-(benzylcycloalkyl)carboxamide and thiocarboxamide derivatives |
| EP2218717A1 (en) | 2009-02-17 | 2010-08-18 | Bayer CropScience AG | Fungicidal N-((HET)Arylethyl)thiocarboxamide derivatives |
| WO2010094728A1 (en) | 2009-02-19 | 2010-08-26 | Bayer Cropscience Ag | Pesticide composition comprising a tetrazolyloxime derivative and a fungicide or an insecticide active substance |
| EP2223602A1 (en) | 2009-02-23 | 2010-09-01 | Bayer CropScience AG | Method for improved utilisation of the production potential of genetically modified plants |
| US8846568B2 (en) | 2009-03-25 | 2014-09-30 | Bayer Cropscience Ag | Active compound combinations having insecticidal and acaricidal properties |
| US8828906B2 (en) | 2009-03-25 | 2014-09-09 | Bayer Cropscience Ag | Active compound combinations having insecticidal and acaricidal properties |
| US8828907B2 (en) | 2009-03-25 | 2014-09-09 | Bayer Cropscience Ag | Active ingredient combinations having insecticidal and acaricidal properties |
| US8846567B2 (en) | 2009-03-25 | 2014-09-30 | Bayer Cropscience Ag | Active compound combinations having insecticidal and acaricidal properties |
| US9012360B2 (en) | 2009-03-25 | 2015-04-21 | Bayer Intellectual Property Gmbh | Synergistic combinations of active ingredients |
| EP2232995A1 (en) | 2009-03-25 | 2010-09-29 | Bayer CropScience AG | Method for improved utilisation of the production potential of transgenic plants |
| EP2239331A1 (en) | 2009-04-07 | 2010-10-13 | Bayer CropScience AG | Method for improved utilization of the production potential of transgenic plants |
| US8835657B2 (en) | 2009-05-06 | 2014-09-16 | Bayer Cropscience Ag | Cyclopentanedione compounds and their use as insecticides, acaricides and/or fungicides |
| EP2251331A1 (en) | 2009-05-15 | 2010-11-17 | Bayer CropScience AG | Fungicide pyrazole carboxamides derivatives |
| EP3000809A1 (en) | 2009-05-15 | 2016-03-30 | Bayer Intellectual Property GmbH | Fungicide pyrazole carboxamides derivatives |
| EP2255626A1 (en) | 2009-05-27 | 2010-12-01 | Bayer CropScience AG | Use of succinate dehydrogenase inhibitors to increase resistance of plants or parts of plants to abiotic stress |
| US9877482B2 (en) | 2009-06-02 | 2018-01-30 | Bayer Intellectual Property Gmbh | Use of succinate dehydrogenase inhibitors for controlling Sclerotinia ssp |
| US9232794B2 (en) | 2009-06-02 | 2016-01-12 | Bayer Intellectual Property Gmbh | Use of succinate dehydrogenase inhibitors for controlling Sclerotinia ssp |
| WO2011015524A2 (en) | 2009-08-03 | 2011-02-10 | Bayer Cropscience Ag | Fungicide heterocycles derivatives |
| EP2292094A1 (en) | 2009-09-02 | 2011-03-09 | Bayer CropScience AG | Active compound combinations |
| WO2011035834A1 (en) | 2009-09-02 | 2011-03-31 | Bayer Cropscience Ag | Active compound combinations |
| EP2343280A1 (en) | 2009-12-10 | 2011-07-13 | Bayer CropScience AG | Fungicide quinoline derivatives |
| WO2011080255A2 (en) | 2009-12-28 | 2011-07-07 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2011080254A2 (en) | 2009-12-28 | 2011-07-07 | Bayer Cropscience Ag | Fungicide hydroximoyl-heterocycles derivatives |
| WO2011080256A1 (en) | 2009-12-28 | 2011-07-07 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| US8722072B2 (en) | 2010-01-22 | 2014-05-13 | Bayer Intellectual Property Gmbh | Acaricidal and/or insecticidal active ingredient combinations |
| WO2011089071A2 (en) | 2010-01-22 | 2011-07-28 | Bayer Cropscience Ag | Acaricide and/or insecticide active substance combinations |
| WO2011107504A1 (en) | 2010-03-04 | 2011-09-09 | Bayer Cropscience Ag | Fluoroalkyl-substituted 2-amidobenzimidazoles and the use thereof for boosting stress tolerance in plants |
| WO2011124554A2 (en) | 2010-04-06 | 2011-10-13 | Bayer Cropscience Ag | Use of 4-phenylbutyric acid and/or the salts thereof for enhancing the stress tolerance of plants |
| WO2011124553A2 (en) | 2010-04-09 | 2011-10-13 | Bayer Cropscience Ag | Use of derivatives of the (1-cyanocyclopropyl)phenylphosphinic acid, the esters thereof and/or the salts thereof for enhancing the tolerance of plants to abiotic stress |
| WO2011134912A1 (en) | 2010-04-28 | 2011-11-03 | Bayer Cropscience Ag | Fungicide hydroximoyl-heterocycles derivatives |
| WO2011134913A1 (en) | 2010-04-28 | 2011-11-03 | Bayer Cropscience Ag | Fungicide hydroximoyl-heterocycles derivatives |
| WO2011134911A2 (en) | 2010-04-28 | 2011-11-03 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2011151368A2 (en) | 2010-06-03 | 2011-12-08 | Bayer Cropscience Ag | Fungicide n-[(trisubstitutedsilyl)methyl]-carboxamide derivatives |
| WO2011151369A1 (en) | 2010-06-03 | 2011-12-08 | Bayer Cropscience Ag | N-[(het)arylethyl)] pyrazole(thio)carboxamides and their heterosubstituted analogues |
| WO2011151370A1 (en) | 2010-06-03 | 2011-12-08 | Bayer Cropscience Ag | N-[(het)arylalkyl)] pyrazole (thio)carboxamides and their heterosubstituted analogues |
| WO2011154158A1 (en) | 2010-06-09 | 2011-12-15 | Bayer Bioscience N.V. | Methods and means to modify a plant genome at a nucleotide sequence commonly used in plant genome engineering |
| WO2011154159A1 (en) | 2010-06-09 | 2011-12-15 | Bayer Bioscience N.V. | Methods and means to modify a plant genome at a nucleotide sequence commonly used in plant genome engineering |
| WO2012010579A2 (en) | 2010-07-20 | 2012-01-26 | Bayer Cropscience Ag | Benzocycloalkenes as antifungal agents |
| WO2012028578A1 (en) | 2010-09-03 | 2012-03-08 | Bayer Cropscience Ag | Substituted fused pyrimidinones and dihydropyrimidinones |
| WO2012038476A1 (en) | 2010-09-22 | 2012-03-29 | Bayer Cropscience Ag | Use of active ingredients for controlling nematodes in nematode-resistant crops |
| WO2012038480A2 (en) | 2010-09-22 | 2012-03-29 | Bayer Cropscience Ag | Use of biological or chemical control agents for controlling insects and nematodes in resistant crops |
| WO2012045798A1 (en) | 2010-10-07 | 2012-04-12 | Bayer Cropscience Ag | Fungicide composition comprising a tetrazolyloxime derivative and a thiazolylpiperidine derivative |
| WO2012052490A1 (en) | 2010-10-21 | 2012-04-26 | Bayer Cropscience Ag | N-benzyl heterocyclic carboxamides |
| WO2012052489A1 (en) | 2010-10-21 | 2012-04-26 | Bayer Cropscience Ag | 1-(heterocyclic carbonyl) piperidines |
| WO2012059497A1 (en) | 2010-11-02 | 2012-05-10 | Bayer Cropscience Ag | N-hetarylmethyl pyrazolylcarboxamides |
| US9206137B2 (en) | 2010-11-15 | 2015-12-08 | Bayer Intellectual Property Gmbh | N-Aryl pyrazole(thio)carboxamides |
| WO2012065947A1 (en) | 2010-11-15 | 2012-05-24 | Bayer Cropscience Ag | 5-halogenopyrazolecarboxamides |
| WO2012065945A1 (en) | 2010-11-15 | 2012-05-24 | Bayer Cropscience Ag | 5-halogenopyrazole(thio)carboxamides |
| WO2012065944A1 (en) | 2010-11-15 | 2012-05-24 | Bayer Cropscience Ag | N-aryl pyrazole(thio)carboxamides |
| EP3103340A1 (en) | 2010-12-01 | 2016-12-14 | Bayer Intellectual Property GmbH | Agent combinations comprising pyridylethyl benzamides and other agents |
| EP3103339A1 (en) | 2010-12-01 | 2016-12-14 | Bayer Intellectual Property GmbH | Agent combinations comprising pyridylethyl benzamides and other agents |
| EP3103338A1 (en) | 2010-12-01 | 2016-12-14 | Bayer Intellectual Property GmbH | Agent combinations comprising pyridylethyl benzamides and other agents |
| EP3103334A1 (en) | 2010-12-01 | 2016-12-14 | Bayer Intellectual Property GmbH | Agent combinations comprising pyridylethyl benzamides and other agents |
| EP3092900A1 (en) | 2010-12-01 | 2016-11-16 | Bayer Intellectual Property GmbH | Active ingredient combinations comprising pyridylethylbenzamides and other active ingredients |
| EP2460407A1 (en) | 2010-12-01 | 2012-06-06 | Bayer CropScience AG | Agent combinations comprising pyridylethyl benzamides and other agents |
| EP2460406A1 (en) | 2010-12-01 | 2012-06-06 | Bayer CropScience AG | Use of fluopyram for controlling nematodes in nematode resistant crops |
| WO2012072660A1 (en) | 2010-12-01 | 2012-06-07 | Bayer Cropscience Ag | Use of fluopyram for controlling nematodes in crops and for increasing yield |
| WO2012072696A1 (en) | 2010-12-01 | 2012-06-07 | Bayer Cropscience Ag | Active ingredient combinations comprising pyridylethylbenzamides and other active ingredients |
| EP2474542A1 (en) | 2010-12-29 | 2012-07-11 | Bayer CropScience AG | Fungicide hydroximoyl-tetrazole derivatives |
| WO2012089757A1 (en) | 2010-12-29 | 2012-07-05 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2012089721A1 (en) | 2010-12-30 | 2012-07-05 | Bayer Cropscience Ag | Use of substituted spirocyclic sulfonamidocarboxylic acids, carboxylic esters thereof, carboxamides thereof and carbonitriles thereof or salts thereof for enhancement of stress tolerance in plants |
| WO2012089722A2 (en) | 2010-12-30 | 2012-07-05 | Bayer Cropscience Ag | Use of open-chain carboxylic acids, carbonic esters, carboxamides and carbonitriles of aryl, heteroaryl and benzylsulfonamide or the salts thereof for improving the stress tolerance in plants |
| EP2494867A1 (en) | 2011-03-01 | 2012-09-05 | Bayer CropScience AG | Halogen-substituted compounds in combination with fungicides |
| WO2012120105A1 (en) | 2011-03-10 | 2012-09-13 | Bayer Cropscience Ag | Use of lipochito-oligosaccharide compounds for safeguarding seed safety of treated seeds |
| WO2012123434A1 (en) | 2011-03-14 | 2012-09-20 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| WO2012136581A1 (en) | 2011-04-08 | 2012-10-11 | Bayer Cropscience Ag | Fungicide hydroximoyl-tetrazole derivatives |
| EP2511255A1 (en) | 2011-04-15 | 2012-10-17 | Bayer CropScience AG | Substituted prop-2-in-1-ol and prop-2-en-1-ol derivatives |
| WO2012139891A1 (en) | 2011-04-15 | 2012-10-18 | Bayer Cropscience Ag | Substituted vinyl and alkinyl cyclohexenols as active agents against abiotic stress in plants |
| WO2012139890A1 (en) | 2011-04-15 | 2012-10-18 | Bayer Cropscience Ag | Substituted 5-(cyclohex-2-en-1-yl)-penta-2,4-dienes and 5-(cyclohex-2-en-1-yl)-pent-2-en-4-ines as active agents against abiotic stress in plants |
| WO2012139892A1 (en) | 2011-04-15 | 2012-10-18 | Bayer Cropscience Ag | Substituted 5-(bicyclo[4.1.0]hept-3-en-2-yl)-penta-2,4-dienes and 5-(bicyclo[4.1.0]hept-3-en-2-yl)-pent-2-ene-4-ines as active agents against abiotic stress in plants |
| EP2997825A1 (en) | 2011-04-22 | 2016-03-23 | Bayer Intellectual Property GmbH | Active compound combinations comprising a (thio)carboxamide derivative and a fungicidal compound |
| WO2012168124A1 (en) | 2011-06-06 | 2012-12-13 | Bayer Cropscience Nv | Methods and means to modify a plant genome at a preselected site |
| WO2013004652A1 (en) | 2011-07-04 | 2013-01-10 | Bayer Intellectual Property Gmbh | Use of substituted isoquinolinones, isoquinolindiones, isoquinolintriones and dihydroisoquinolinones or in each case salts thereof as active agents against abiotic stress in plants |
| WO2013020985A1 (en) | 2011-08-10 | 2013-02-14 | Bayer Intellectual Property Gmbh | Active compound combinations comprising specific tetramic acid derivatives |
| US9265252B2 (en) | 2011-08-10 | 2016-02-23 | Bayer Intellectual Property Gmbh | Active compound combinations comprising specific tetramic acid derivatives |
| WO2013026740A2 (en) | 2011-08-22 | 2013-02-28 | Bayer Cropscience Nv | Methods and means to modify a plant genome |
| US9670496B2 (en) | 2011-08-22 | 2017-06-06 | Bayer Cropscience N.V. | Methods and means to modify a plant genome |
| US10538774B2 (en) | 2011-08-22 | 2020-01-21 | Basf Agricultural Solutions Seed, Us Llc | Methods and means to modify a plant genome |
| WO2013026836A1 (en) | 2011-08-22 | 2013-02-28 | Bayer Intellectual Property Gmbh | Fungicide hydroximoyl-tetrazole derivatives |
| EP2561759A1 (en) | 2011-08-26 | 2013-02-27 | Bayer Cropscience AG | Fluoroalkyl-substituted 2-amidobenzimidazoles and their effect on plant growth |
| WO2013034621A1 (en) | 2011-09-09 | 2013-03-14 | Bayer Intellectual Property Gmbh | Acyl-homoserine lactone derivatives for improving plant yield |
| WO2013037717A1 (en) | 2011-09-12 | 2013-03-21 | Bayer Intellectual Property Gmbh | Fungicidal 4-substituted-3-{phenyl[(heterocyclylmethoxy)imino]methyl}-1,2,4-oxadizol-5(4h)-one derivatives |
| WO2013037956A1 (en) | 2011-09-16 | 2013-03-21 | Bayer Intellectual Property Gmbh | Use of 5-phenyl- or 5-benzyl-2 isoxazoline-3 carboxylates for improving plant yield |
| WO2013037955A1 (en) | 2011-09-16 | 2013-03-21 | Bayer Intellectual Property Gmbh | Use of acylsulfonamides for improving plant yield |
| WO2013037958A1 (en) | 2011-09-16 | 2013-03-21 | Bayer Intellectual Property Gmbh | Use of phenylpyrazolin-3-carboxylates for improving plant yield |
| WO2013041602A1 (en) | 2011-09-23 | 2013-03-28 | Bayer Intellectual Property Gmbh | Use of 4-substituted 1-phenyl-pyrazole-3-carboxylic-acid derivatives as agents against abiotic plant stress |
| WO2013050410A1 (en) | 2011-10-04 | 2013-04-11 | Bayer Intellectual Property Gmbh | RNAi FOR THE CONTROL OF FUNGI AND OOMYCETES BY INHIBITING SACCHAROPINE DEHYDROGENASE GENE |
| WO2013050324A1 (en) | 2011-10-06 | 2013-04-11 | Bayer Intellectual Property Gmbh | Combination, containing 4-phenylbutyric acid (4-pba) or a salt thereof (component (a)) and one or more selected additional agronomically active compounds (component(s) (b)), that reduces abiotic plant stress |
| WO2013075817A1 (en) | 2011-11-21 | 2013-05-30 | Bayer Intellectual Property Gmbh | Fungicide n-[(trisubstitutedsilyl)methyl]-carboxamide derivatives |
| WO2013079566A2 (en) | 2011-11-30 | 2013-06-06 | Bayer Intellectual Property Gmbh | Fungicidal n-bicycloalkyl and n-tricycloalkyl (thio)carboxamide derivatives |
| WO2013092519A1 (en) | 2011-12-19 | 2013-06-27 | Bayer Cropscience Ag | Use of anthranilic acid diamide derivatives for pest control in transgenic crops |
| WO2013098146A1 (en) | 2011-12-29 | 2013-07-04 | Bayer Intellectual Property Gmbh | Fungicidal 3-[(1,3-thiazol-4-ylmethoxyimino)(phenyl)methyl]-2-substituted-1,2,4-oxadiazol-5(2h)-one derivatives |
| WO2013098147A1 (en) | 2011-12-29 | 2013-07-04 | Bayer Intellectual Property Gmbh | Fungicidal 3-[(pyridin-2-ylmethoxyimino)(phenyl)methyl]-2-substituted-1,2,4-oxadiazol-5(2h)-one derivatives |
| WO2013124275A1 (en) | 2012-02-22 | 2013-08-29 | Bayer Cropscience Ag | Use of succinate dehydrogenase inhibitors (sdhis) for controlling wood diseases in grape. |
| WO2013127704A1 (en) | 2012-02-27 | 2013-09-06 | Bayer Intellectual Property Gmbh | Active compound combinations containing a thiazoylisoxazoline and a fungicide |
| WO2013139949A1 (en) | 2012-03-23 | 2013-09-26 | Bayer Intellectual Property Gmbh | Compositions comprising a strigolactame compound for enhanced plant growth and yield |
| WO2013153143A1 (en) | 2012-04-12 | 2013-10-17 | Bayer Cropscience Ag | N-acyl- 2 - (cyclo) alkylpyrrolidines and piperidines useful as fungicides |
| WO2013156560A1 (en) | 2012-04-20 | 2013-10-24 | Bayer Cropscience Ag | N-cycloalkyl-n-[(trisubstitutedsilylphenyl)methylene]-(thio)carboxamide derivatives |
| WO2013156559A1 (en) | 2012-04-20 | 2013-10-24 | Bayer Cropscience Ag | N-cycloalkyl-n-[(heterocyclylphenyl)methylene]-(thio)carboxamide derivatives |
| US11518997B2 (en) | 2012-04-23 | 2022-12-06 | BASF Agricultural Solutions Seed US LLC | Targeted genome engineering in plants |
| WO2013160230A1 (en) | 2012-04-23 | 2013-10-31 | Bayer Cropscience Nv | Targeted genome engineering in plants |
| EP2662361A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazol indanyl carboxamides |
| EP2662360A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole indanyl carboxamides |
| EP2662362A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazole indanyl carboxamides |
| EP2662363A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole biphenylcarboxamides |
| WO2013167545A1 (en) | 2012-05-09 | 2013-11-14 | Bayer Cropscience Ag | Pyrazole indanyl carboxamides |
| WO2013167544A1 (en) | 2012-05-09 | 2013-11-14 | Bayer Cropscience Ag | 5-halogenopyrazole indanyl carboxamides |
| EP2662370A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | 5-Halogenopyrazole benzofuranyl carboxamides |
| EP2662364A1 (en) | 2012-05-09 | 2013-11-13 | Bayer CropScience AG | Pyrazole tetrahydronaphthyl carboxamides |
| WO2013174836A1 (en) | 2012-05-22 | 2013-11-28 | Bayer Cropscience Ag | Active compounds combinations comprising a lipo-chitooligosaccharide derivative and a nematicide, insecticidal or fungicidal compound |
| WO2014009322A1 (en) | 2012-07-11 | 2014-01-16 | Bayer Cropscience Ag | Use of fungicidal combinations for increasing the tolerance of a plant towards abiotic stress |
| WO2014037340A1 (en) | 2012-09-05 | 2014-03-13 | Bayer Cropscience Ag | Use of substituted 2-amidobenzimidazoles, 2-amidobenzoxazoles and 2-amidobenzothiazoles or salts thereof as active substances against abiotic plant stress |
| WO2014060518A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Method of plant growth promotion using carboxamide derivatives |
| WO2014060502A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Active compound combinations comprising carboxamide derivatives |
| WO2014060520A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Method for treating plants against fungi resistant to fungicides using carboxamide or thiocarboxamide derivatives |
| WO2014060519A1 (en) | 2012-10-19 | 2014-04-24 | Bayer Cropscience Ag | Method for enhancing tolerance to abiotic stress in plants using carboxamide or thiocarboxamide derivatives |
| WO2014079957A1 (en) | 2012-11-23 | 2014-05-30 | Bayer Cropscience Ag | Selective inhibition of ethylene signal transduction |
| WO2014079789A1 (en) | 2012-11-23 | 2014-05-30 | Bayer Cropscience Ag | Active compound combinations |
| EP2735231A1 (en) | 2012-11-23 | 2014-05-28 | Bayer CropScience AG | Active compound combinations |
| WO2014083089A1 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Ternary fungicidal and pesticidal mixtures |
| WO2014083033A1 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropsience Ag | Binary fungicidal or pesticidal mixture |
| WO2014082950A1 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Ternary fungicidal mixtures |
| WO2014083088A2 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Binary fungicidal mixtures |
| WO2014083031A2 (en) | 2012-11-30 | 2014-06-05 | Bayer Cropscience Ag | Binary pesticidal and fungicidal mixtures |
| EP2740720A1 (en) | 2012-12-05 | 2014-06-11 | Bayer CropScience AG | Substituted bicyclic and tricyclic pent-2-en-4-inic acid derivatives and their use for enhancing the stress tolerance in plants |
| EP2740356A1 (en) | 2012-12-05 | 2014-06-11 | Bayer CropScience AG | Substituted (2Z)-5(1-Hydroxycyclohexyl)pent-2-en-4-inic acid derivatives |
| WO2014086751A1 (en) | 2012-12-05 | 2014-06-12 | Bayer Cropscience Ag | Use of substituted 1-(aryl ethynyl)-, 1-(heteroaryl ethynyl)-, 1-(heterocyclyl ethynyl)- and 1-(cyloalkenyl ethynyl)-cyclohexanols as active agents against abiotic plant stress |
| WO2014090765A1 (en) | 2012-12-12 | 2014-06-19 | Bayer Cropscience Ag | Use of 1-[2-fluoro-4-methyl-5-(2,2,2-trifluoroethylsulfinyl)phenyl]-5-amino-3-trifluoromethyl)-1 h-1,2,4 tfia zole for controlling nematodes in nematode-resistant crops |
| WO2014095826A1 (en) | 2012-12-18 | 2014-06-26 | Bayer Cropscience Ag | Binary fungicidal and bactericidal combinations |
| WO2014095677A1 (en) | 2012-12-19 | 2014-06-26 | Bayer Cropscience Ag | Difluoromethyl-nicotinic- tetrahydronaphtyl carboxamides |
| WO2014135608A1 (en) | 2013-03-07 | 2014-09-12 | Bayer Cropscience Ag | Fungicidal 3-{phenyl[(heterocyclylmethoxy)imino]methyl}-heterocycle derivatives |
| WO2014161821A1 (en) | 2013-04-02 | 2014-10-09 | Bayer Cropscience Nv | Targeted genome engineering in eukaryotes |
| WO2014167008A1 (en) | 2013-04-12 | 2014-10-16 | Bayer Cropscience Ag | Novel triazolinthione derivatives |
| WO2014167009A1 (en) | 2013-04-12 | 2014-10-16 | Bayer Cropscience Ag | Novel triazole derivatives |
| WO2014170364A1 (en) | 2013-04-19 | 2014-10-23 | Bayer Cropscience Ag | Binary insecticidal or pesticidal mixture |
| WO2014170345A2 (en) | 2013-04-19 | 2014-10-23 | Bayer Cropscience Ag | Method for improved utilization of the production potential of transgenic plants |
| WO2014177514A1 (en) | 2013-04-30 | 2014-11-06 | Bayer Cropscience Ag | Nematicidal n-substituted phenethylcarboxamides |
| WO2014177582A1 (en) | 2013-04-30 | 2014-11-06 | Bayer Cropscience Ag | N-(2-fluoro-2-phenethyl)carboxamides as nematicides and endoparasiticides |
| WO2014206953A1 (en) | 2013-06-26 | 2014-12-31 | Bayer Cropscience Ag | N-cycloalkyl-n-[(bicyclylphenyl)methylene]-(thio)carboxamide derivatives |
| WO2015004040A1 (en) | 2013-07-09 | 2015-01-15 | Bayer Cropscience Ag | Use of selected pyridone carboxamides or salts thereof as active substances against abiotic plant stress |
| WO2015082587A1 (en) | 2013-12-05 | 2015-06-11 | Bayer Cropscience Ag | N-cycloalkyl-n-{[2-(1-substitutedcycloalkyl)phenyl]methylene}-(thio)carboxamide derivatives |
| WO2015082586A1 (en) | 2013-12-05 | 2015-06-11 | Bayer Cropscience Ag | N-cycloalkyl-n-{[2-(1-substitutedcycloalkyl)phenyl]methylene}-(thio)carboxamide derivatives |
| WO2016012362A1 (en) | 2014-07-22 | 2016-01-28 | Bayer Cropscience Aktiengesellschaft | Substituted cyano cycloalkyl penta-2,4-dienes, cyano cycloalkyl pent-2-en-4-ynes, cyano heterocyclyl penta-2,4-dienes and cyano heterocyclyl pent-2-en-4-ynes as active substances against abiotic plant stress |
| WO2016096942A1 (en) | 2014-12-18 | 2016-06-23 | Bayer Cropscience Aktiengesellschaft | Use of selected pyridone carboxamides or salts thereof as active substances against abiotic plant stress |
| WO2016166077A1 (en) | 2015-04-13 | 2016-10-20 | Bayer Cropscience Aktiengesellschaft | N-cycloalkyl-n-(biheterocyclyethylene)-(thio)carboxamide derivatives |
| US11180751B2 (en) | 2015-06-18 | 2021-11-23 | The Broad Institute, Inc. | CRISPR enzymes and systems |
| WO2018019676A1 (en) | 2016-07-29 | 2018-02-01 | Bayer Cropscience Aktiengesellschaft | Active compound combinations and methods to protect the propagation material of plants |
| WO2018054829A1 (en) | 2016-09-22 | 2018-03-29 | Bayer Cropscience Aktiengesellschaft | Novel triazole derivatives and their use as fungicides |
| WO2018054832A1 (en) | 2016-09-22 | 2018-03-29 | Bayer Cropscience Aktiengesellschaft | Novel triazole derivatives |
| WO2018054911A1 (en) | 2016-09-23 | 2018-03-29 | Bayer Cropscience Nv | Targeted genome optimization in plants |
| US12499971B2 (en) | 2016-09-28 | 2025-12-16 | The Broad Institute, Inc. | Systematic screening and mapping of regulatory elements in non-coding genomic regions, methods, compositions, and applications thereof |
| WO2018077711A2 (en) | 2016-10-26 | 2018-05-03 | Bayer Cropscience Aktiengesellschaft | Use of pyraziflumid for controlling sclerotinia spp in seed treatment applications |
| WO2018104392A1 (en) | 2016-12-08 | 2018-06-14 | Bayer Cropscience Aktiengesellschaft | Use of insecticides for controlling wireworms |
| WO2018108627A1 (en) | 2016-12-12 | 2018-06-21 | Bayer Cropscience Aktiengesellschaft | Use of substituted indolinylmethyl sulfonamides, or the salts thereof for increasing the stress tolerance of plants |
| EP3332645A1 (en) | 2016-12-12 | 2018-06-13 | Bayer Cropscience AG | Use of substituted pyrimidine diones or their salts as agents to combat abiotic plant stress |
| US11591601B2 (en) | 2017-05-05 | 2023-02-28 | The Broad Institute, Inc. | Methods for identification and modification of lncRNA associated with target genotypes and phenotypes |
| US12297436B2 (en) | 2017-05-18 | 2025-05-13 | The Broad Institute, Inc. | Systems, methods, and compositions for targeted nucleic acid editing |
| WO2019025153A1 (en) | 2017-07-31 | 2019-02-07 | Bayer Cropscience Aktiengesellschaft | USE OF SUBSTITUTED N-SULFONYL-N'-ARYLDIAMINOALKANES AND N-SULFONYL-N'-HETEROARYL DIAMINOALKANES OR THEIR SALTS TO INCREASE STRESSTOLERANCE IN PLANTS |
| WO2019060746A1 (en) | 2017-09-21 | 2019-03-28 | The Broad Institute, Inc. | Systems, methods, and compositions for targeted nucleic acid editing |
| WO2019126709A1 (en) | 2017-12-22 | 2019-06-27 | The Broad Institute, Inc. | Cas12b systems, methods, and compositions for targeted dna base editing |
| US10968257B2 (en) | 2018-04-03 | 2021-04-06 | The Broad Institute, Inc. | Target recognition motifs and uses thereof |
| US11999767B2 (en) | 2018-04-03 | 2024-06-04 | The Broad Institute, Inc. | Target recognition motifs and uses thereof |
| WO2019233863A1 (en) | 2018-06-04 | 2019-12-12 | Bayer Aktiengesellschaft | Herbicidally active bicyclic benzoylpyrazoles |
| WO2020131862A1 (en) | 2018-12-17 | 2020-06-25 | The Broad Institute, Inc. | Crispr-associated transposase systems and methods of use thereof |
Also Published As
| Publication number | Publication date |
|---|---|
| AU7647000A (en) | 2001-03-19 |
| AR026151A1 (en) | 2003-01-29 |
| WO2001014569A3 (en) | 2001-10-11 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| WO2001014569A2 (en) | Increasing the polysaccharide content in plants | |
| EP1038014B1 (en) | VEGETABLE (GntI) SEQUENCES AND THE USE THEREOF TO OBTAIN PLANTS WITH A REDUCED OR LACK OF N-ACETYLGLUCOSAMINYLTRANSFERASE I (GnTI) ACTIVITY | |
| DE69233380T2 (en) | TRANSGENIC PLANTS WITH CHANGED POLYOL CONTENT | |
| DE69333180T2 (en) | DNA SEQUENCES CODING FOR OLIGOSACCHARIDE TRANSPORTER | |
| EP1009841A1 (en) | Dna sequence coding for a hydroxyphenylpyruvate dioxygenase and overproduction thereof in plants | |
| DE69132758T2 (en) | Plasmids for the production of transgenic plants which have changed their habit and yield | |
| EP1102852A1 (en) | Dna sequence coding for a 1-deoxy-d-xylulose-5-phosphate synthase and the overproduction thereof in plants | |
| DE19502053A1 (en) | Processes and DNA molecules to increase the rate of photosynthesis in plants, as well as plant cells and plants with an increased rate of photosynthesis | |
| EP1212439B1 (en) | Plants having altered amino acid contents and method for the production thereof | |
| DE19752647C1 (en) | Reduction of the chlorophyll content in oil plant seeds | |
| DE19853778C1 (en) | DNA sequences encoding a glutamate / malate translocator, plasmid bacteria, yeast and plants containing this transporter | |
| WO1998006831A1 (en) | Transgenic plant cells and plants with modified acetyl-coa formation | |
| AU719452B2 (en) | Transgenic plant cells and plants with an increased glycolysis rate | |
| EP1070120A1 (en) | Amp deaminase | |
| DE19903493A1 (en) | Overexpression of a DNA sequence coding for a transketolase in plants | |
| WO1998010074A2 (en) | Adenylosuccinate synthetase | |
| DE10313795A1 (en) | Altered PPase expression in sugar beet | |
| DE19632121C2 (en) | Transgenic plant cells and plants with altered acetyl-CoA formation | |
| DE19732926C2 (en) | DNA sequences encoding a glucose-6-phosphate-phosphate translocator, as well as plasmids, bacteria, yeasts and plants containing this transporter | |
| WO2000060101A1 (en) | Metabolic selection markers for plants | |
| EP1210437B1 (en) | Dihydroorotase extracted from plants | |
| EP1198578A2 (en) | PLANT S-ADENOSYLMETHIONIN:Mg-PROTOPORPHYRIN-IX-O-METHYLTRANSFERASE, PLANTS WITH VARIABLE CHLOROPHYLL CONTENTS AND/OR HERBICIDE TOLERANCE, AND METHOD FOR THE PRODUCTION THEREOF | |
| EP1294925A2 (en) | Phosphoribosyl pyrophosphate synthetase 1 as herbicidal target | |
| Lal | Transcriptional and translational regulation of enolase under anaerobic stress in maize | |
| DE10104721A1 (en) | Process for increasing the content of sulfur compounds in plants |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| AK | Designated states |
Kind code of ref document: A2 Designated state(s): AE AG AL AM AT AU AZ BA BB BG BR BY BZ CA CH CN CR CU CZ DE DK DM DZ EE ES FI GB GD GE GH GM HR HU ID IL IN IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MA MD MG MK MN MW MX MZ NO NZ PL PT RO RU SD SE SG SI SK SL TJ TM TR TT TZ UA UG US UZ VN YU ZA ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A2 Designated state(s): GH GM KE LS MW MZ SD SL SZ TZ UG ZW AM AZ BY KG KZ MD RU TJ TM AT BE CH CY DE DK ES FI FR GB GR IE IT LU MC NL PT SE BF BJ CF CG CI CM GA GN GW ML MR NE SN TD TG |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application | ||
| DFPE | Request for preliminary examination filed prior to expiration of 19th month from priority date (pct application filed before 20040101) | ||
| AK | Designated states |
Kind code of ref document: A3 Designated state(s): AE AG AL AM AT AU AZ BA BB BG BR BY BZ CA CH CN CR CU CZ DE DK DM DZ EE ES FI GB GD GE GH GM HR HU ID IL IN IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MA MD MG MK MN MW MX MZ NO NZ PL PT RO RU SD SE SG SI SK SL TJ TM TR TT TZ UA UG US UZ VN YU ZA ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A3 Designated state(s): GH GM KE LS MW MZ SD SL SZ TZ UG ZW AM AZ BY KG KZ MD RU TJ TM AT BE CH CY DE DK ES FI FR GB GR IE IT LU MC NL PT SE BF BJ CF CG CI CM GA GN GW ML MR NE SN TD TG |
|
| REG | Reference to national code |
Ref country code: DE Ref legal event code: 8642 |
|
| 122 | Ep: pct application non-entry in european phase | ||
| NENP | Non-entry into the national phase |
Ref country code: JP |