Abstract
Objective: To examine the expression and function of long non-coding RNA taurine up-regulated 1 (TUG1) in human osteosarcoma cells. Methods: Real-time quantitive PCR was used to detect the transcription level of TUG1 in a series of osteosarcoma cell lines. Knockdown of TUG1 in U2OS cells was carried out by transient transfection of siRNAs. MTT assay was performed to access the cell growth rates. Afterwards, RNA and protein of these cells were extracted to analyze the transfection efficient as well as the expression of other molecules. Results: Compared to the normal cell line, TUG1 exhibited a significant upregulation in osteosarcoma cells. Phenotyping analysis showed the growth-promotion activity of TUG1, since knockdown of TUG1 resulted in declined proliferation. We also found that AKT phosphorylation was impaired after TUG1 was inhibited, suggesting that the AKT pathway was involved in the regulation of TUG1 in U2OS cells. Conclusion: Our data provided evidence that TUG1 was upregulated and acted as a possible oncogene via positively regulating cell proliferation in osteosarcoma cells.
1 Introduction
Long non-coding RNA (LncRNA) is a group of transcribed non-coding RNAs that contain more than 200 nt in length and not encoding any proteins [1,2]. They constitute a major but still poorly characterized part of human transcriptome. They were initially considered as genomic noise. Thanks to the highly thorough sequencing as well as functional technologies in recent years, lncRNAs have been demonstrated to play important roles in basic biological processes [3-5], like DNA replication, gene transcription and epigenetic modification. Recently, we gradually recognized that lncRNAs were closely correlated with human carcinogenesis since they had been proved to be abnormally expressed and act as either oncogenes or tumor suppressors [6-9]. In addition, it is a hot topic to discuss the clinical application of specific lncRNAs as a potential novel class of biomarkers for cancer diagnosis and prognosis [10-12].
Taurine up-regulated 1 (TUG1), a 7.1-kb lncRNA, was a long non-coding RNA that was characterized first two years ago in a paper exploring differential expression of lncRNAs in DNA damage-induced cell death in Hela cells [13]. In the following studies, it was clear that TUG1 was increased in bladder urothelial carcinomas, and promote cell proliferation and apoptosis-inhibition [14]. It exhibited similar high expression and pro-tumorous effects in esophageal squamous cell carcinoma, in which TUG1 had the ability to promote cell migration [15]. However, in other types of cancers, including non-small cell lung cancer [16], TUG1 was reported to be downregulated and inhibits cell proliferation as well as inducing apoptosis. This cancer type-specific activity of TUG1 illustrated the importance of the microenvironment. Although it was already found that in osteosarcoma TUG1 was upregulated [17], the related mechanisms were still lacking. In our study, experiments were designed to access its expression, function and possible signaling pathway that TUG1 regulated.
2 Materials and Methods
2.1 Cell Culture
Human normal osteoblastic cell line hFOB1.19 and human Human normal osteoblastic cell line hFOB1.19 and human osteosarcoma cell line SaoS2, MG63, U2OS and HOS were ordered from the Cell Line Resource Center, Shanghai Institutes for Biological Sciences, the Chinese Academy of Sciences (Shanghai, China) or ATCC (American Type Culture Collection, USA). All these cells lines were cultured in 1640 or DMEM supplemented with 10% fetal bovine serum, penicillin (100units/ml) and streptomycin (100μg/ml) at 37°C in a humidified 5% CO2 atmosphere.
2.2 Reagents and Antibodies
Both the control siRNA and LncRNA TUG1 siRNA were purchased by GenePharma (Shanghai, China). The cells were transfected with mimics or inhibitors using Lipofectamine RNAiMAX as described below. PCNA and β-actin antibodies were purchased from Santa Cruz Biotechnology Inc. Phosphated-Akt and total-Akt antibodies were purchased from Cell Signaling Technology Inc. Phosphated-Akt and total-Akt antibodies were purchased from Cell Signaling Technology Inc.
2.3 Transient Transfection
Transfections were performed using Lipofectamine RNAiMAX Reagent (Invitrogene) as per the manufacturer’s instructions. After 48 hours, cells were harvest for the other experiments.
2.4 Real-Time PCR Analysis
Total RNAs was extracted by TRIZOL from the cell lines of human osteosarcoma. Real-time PCRs were performed using an ABI7900 real-time PCR system (Applied Biosystems, Carlsbad, CA) and the SYBR Premix Ex Taq reagent kit (Takara Bio Inc., Shiga, Japan) using Ct quantization method. Ct value was the number of PCR cycles at which the fluorescence signal exceeded the threshold. ∆Ct was the difference in Ct values between the control (GAPDH) and test targets (LncRNA TUG1). ∆∆Ct was the difference in ∆Ct values between the experimental group and paired control group, which represents the fold change in long non-coding RNA expression. The primers change in long non-coding RNA expression. The primers were designed by Primer premier 5.0, and the sequences were as follows:
TUG1
Forward 5’-CTGAAGAAAGGCAA CATC-3’;
Reverse 5’-GTAGGCTACTACAGGATTTG-3’;
GAPDH
Forward 5’-GTCAACGGATTTGGTCTGTATT-3’;
Reverse 5’-AGTCTTCTGGGTGGCAGTGAT-3’.
2.5 Western Blot
For western blot analysis, in brief, the cell lysate was run on SDS-PAGE in 9% acrylamide gels and transferred onto nitrocellulose membranes. After blocking, blots were incubated with mAb. β-actin was used to normalise protein loading. A total of 30μg of cell lysate was loaded in each lane for western blot analysis.
2.6 MTT Assay
Cellular growth ability was determined by the MTT assay. In brief, 3×103 U2OS cells/well were plated into 96-well plates. At the indicated time point, 5μl MTT solution (5mg/ ml) was added into each well and incubated for 2h at room temperature (RT). Then the reaction was terminated by 100μl DMSO and the absorbance at 590nm was measured on a microplate-reader.
2.7 Statistic Analysis
Data are presented as mean ± sd of at least triplicate experiments. Statistical analyses were performed by using SPSS version 18.0 (SPSS, Chicago, IL) and GraphPad Prism Software (GraphPad Software, Inc., San Diego, CA). For all statistical analyses, a P value of < 0.05 was considered statistically significant.
3 Results
Increased expression of TUG1 in osteosarcoma cell lines
Firstly, we applied real-time PCR to quantify the expression of TUG1 in osteosarcoma cell lines. As shown in Figure 1A, when compared to the normal hFOB1.19 cell line, the expression of TUG1 was significantly upregulated in a panel of osteosarcoma cells, including SaoS2, MG63, U2OS and HOS cell lines. This finding was consistent with the previous conclusion that supported its upregulation in clinical osteosarcoma tissue samples [17].
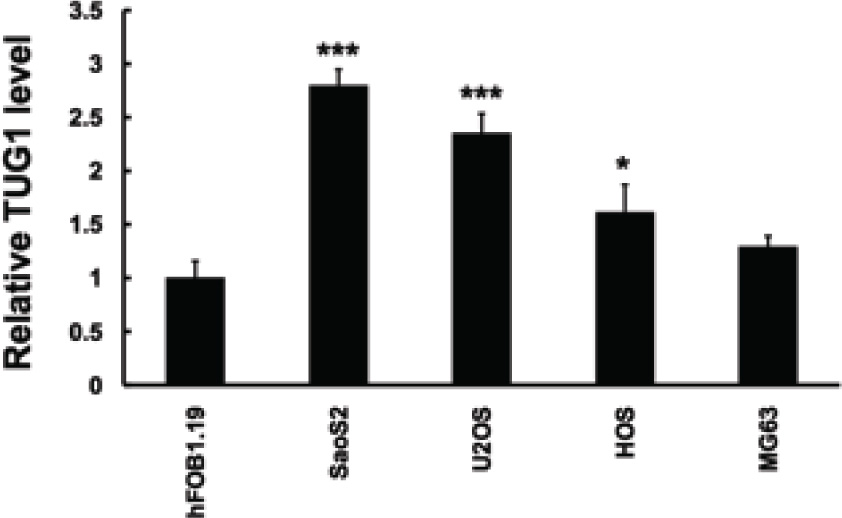
LncRNA TUG1 is upregulated in osteosarcoma cell line. QRT-PCR analysis performed to detect expression of LncRNA TUG1 expression in human normal osteoblastic cell line hF081.19 and human osteosarcoma cell lines. LncRNA TUG1 expression levels were calculated by the 2-∆CT method and normalized to GAPDH. ∆Ct is the difference in Ct values between LncRNA and GAPDH. The plot shows the TUG1 expression was elevated.
3.1 TUG1 has a growth-promotion activity in osteosarcoma
Afterwards, TUG1 siRNA was designed and transiently transfected into the U2OS cells to knockdown the endogenous expression of TUG1. The knockdown efficiency was revealed to be effective, as approximately 80% of TUG1 expression was knockdown (Figure 2A). The MTT assay was performed to visualize the proliferation of cells. As shown in Figure 2B, after TUG1 was inhibited, the growth rate of U2OS cells had decreased compared with the cells in the control group. These results confirmed the previous conclusion that regards TUG1 as a potential oncogene via regulating cell proliferation.
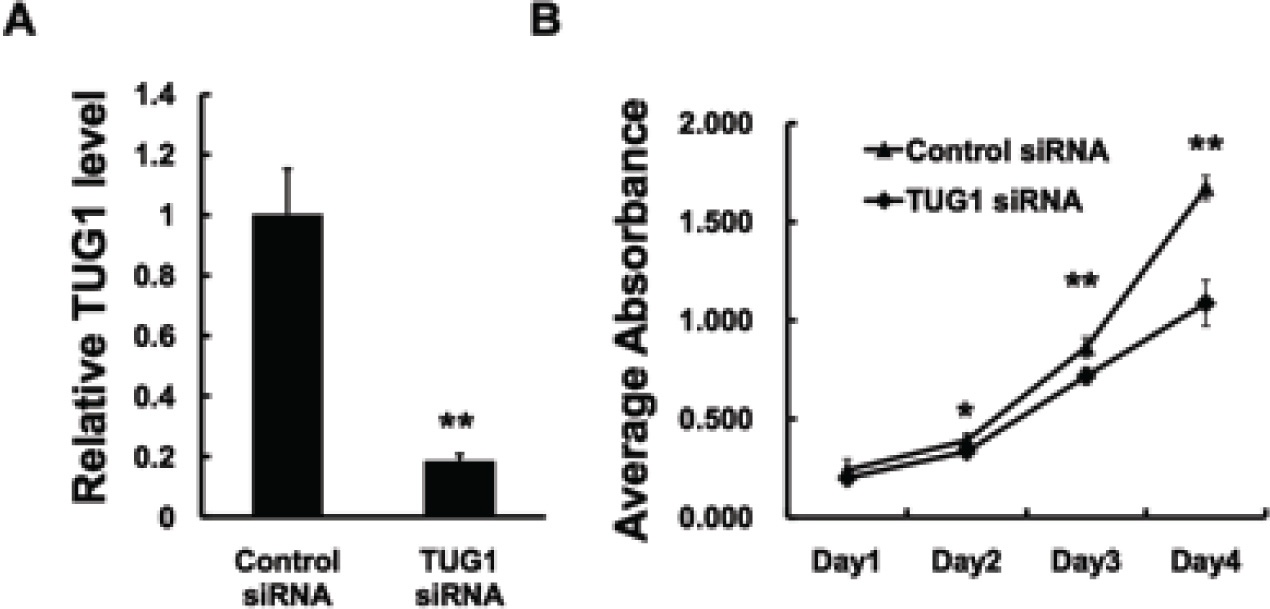
TUG1 promotes tumor proliferation in U2OS cells. (A) U2OS cells were transfected with control siRNA or lncRNA TUG1 siRNA. In 48h, the cells were harvested for examining knockdown effect. (B) U2OS cells were transfected with control siRNA or lncRNA TUG1 siRNA. In 24h, cells were trypsinized and seeded into four 96-well plates at a density of 3×103 cells for MTT assay as described under Materials and Methods. Data are expressed as the mean ± s.d. of the experiments performed in triplicate. *P < 0.05, **P < 0.01.
3.2 TUG1 modulated the AKT signaling
To further understand the possible mechanisms that govern the pro-growth function of TUG1 in U2OS cells, we detected proliferation-related molecules including proliferating cell nuclear antigen (PCNA) and the well known AKT signaling pathway. As shown in Figure 3, after TUG1 was knocked down, the expression of PCNA was decreased, reflecting the growth inhibition at the molecule level. In addition, we found that the phosphor-AKT (at Ser 473) was also decreased in the TUG1-siRNA group, whereas the total AKT expression was unchanged. Since the phosphorylated AKT was an important transducer of AKT signaling pathway, this result suggested that the oncogenic activity of TUG1 might act through this pathway at least partially.
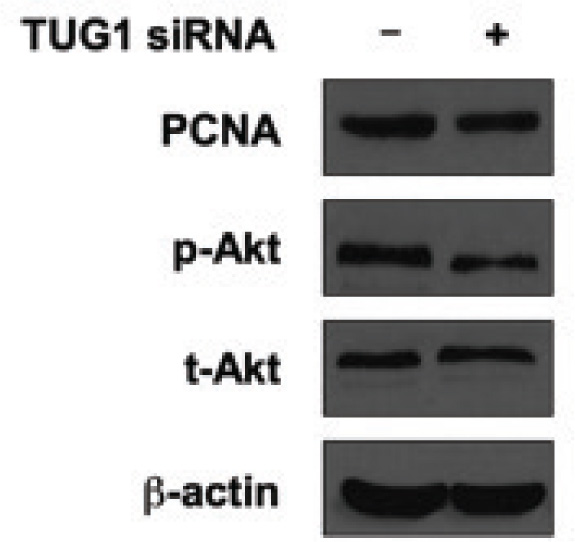
Knockdown of TUG1 reduces survival markers in vitro. U2OS cells were transfected with control siRNA or lncRNA TUG1 siRNA. Forty-eight hours later, cells were harvested for western blotting. Protein level of PCNA, p-Akt (S473) and t-Akt were tested. β-actin was used as the loading control in western blotting. The results showed that inhibited expression of TUG1 suppressed cell proliferation marker PCNA, and also suppressed cell survival marker p-Akt.
4 Discussion
Osteosarcoma is the most common primary bone malignant tumor in children and young adults. Although it has small incidence, the high degree of malignancy makes it a second leading cause of cancer-related death in this age group [18]. The 5-year survival rate is about 65% for patients with localized osteosarcoma, however, it decreases to only about 20% for these cases with metastasis [19-20].
Moreover, objective and reliable diagnostic biomarkers and effective targeted therapeutic agents are also lacking. Thus, further elucidating the underlying mechanisms of osteosarcoma is urgently required. According to the recent studies, lncRNAs emerge as a new frontier of translational research from molecular biology to cancer in the clinic.
TUG1 has been reported in osteosarcoma to be abnormally expressed and accelerates cell proliferation. However, the precise mechanisms or the specific signaling pathways it involves remain unknown. In the present study, we similarly demonstrated its upregulation in four osteosarcoma cell lines, compared to the normal control cells. In in vitro experiments, we found that after TUG1 was inhibited, the growth rate of U2OS cells was reduced. These findings once again confirmed the former study [17]. Besides, we found that TUG1 might regulate the expression of PCNA, and phosho-AKT (at Ser 473), but the total level of AKT remains unchanged after TUG1 was knocked down. The AKT pathway or PI3K-AKT pathway is a signal transduction pathway that promotes survival and growth in response to extracellular signals. AKT is activated by phosphorylation at several sites. Activated AKT mediates downstream responses, including cell survival, growth, proliferation, cell migration and angiogenesis, by phosphorylating a range of intracellular proteins. Our results indicated that TUG1 might promote cell proliferation in osteosarcoma cells through AKT signaling pathway.
In summary, we demonstrated that TUG1 was increased and promoted cell proliferation in osteosarcoma cells. Mechanically, we proved that AKT signaling was involved in this oncogenic regulation. Further studies need to be carried out to verify its pro-tumor activity and clarify the mechanisms in detail.
Conflict of interest statement
Authors state no conflict of interest.
References
[1] Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155-15910.1038/nrg2521Search in Google Scholar PubMed
[2] Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;20;136(4):629-64110.1016/j.cell.2009.02.006Search in Google Scholar PubMed
[3] Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;8;316(5830):1484-148810.1126/science.1138341Search in Google Scholar PubMed
[4] Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2015;11. pii: S1874-9399(15)00123-12610.1016/j.bbagrm.2015.06.003Search in Google Scholar PubMed PubMed Central
[5] Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;29;87:15-2410.1016/j.addr.2015.05.012Search in Google Scholar PubMed PubMed Central
[6] Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015 Jun 12;3:510.1186/s40591-015-0042-6Search in Google Scholar PubMed PubMed Central
[7] Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed). 2015 ;1;7:94-10810.2741/s427Search in Google Scholar
[8] Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12(1):1-9Search in Google Scholar
[9] Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;28;361(1):13-2110.1016/j.canlet.2015.03.002Search in Google Scholar PubMed
[10] Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;23;6:14510.3389/fgene.2015.00145Search in Google Scholar PubMed PubMed Central
[11] Jin K, Luo G, Xiao Z, Liu Z, Liu C, Ji S, Xu J, Liu L, Long J, Ni Q, Yu X. Noncoding RNAs as potential biomarkers to predict the outcome in pancreatic cancer. Drug Des Devel Ther. 2015;26;9:1247-125510.2147/DDDT.S77597Search in Google Scholar PubMed PubMed Central
[12] Ye LC, Zhu X, Qiu JJ, Xu J, Wei Y. Involvement of long non-coding RNA in colorectal cancer: From benchtop to bedside (Review). Oncol Lett. 2015;9(3):1039-104510.3892/ol.2015.2846Search in Google Scholar
[13] Özgür E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin Exp Med. 2013;13(2):119-12610.1007/s10238-012-0181-xSearch in Google Scholar
[14] Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107(5):555-55910.1002/jso.23264Search in Google Scholar
[15] Xu Y, Wang J, Qiu M, Xu L, Li M, Jiang F, Yin R, Xu L. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol. 2015;36(3):1643-165110.1007/s13277-014-2763-6Search in Google Scholar
[16] Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, De W, Shu YQ, Wang ZX. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014; 22;5:e124310.1038/cddis.2014.201Search in Google Scholar
[17] Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prey. 2013;14(4):2311-231510.7314/APJCP.2013.14.4.2311Search in Google Scholar
[18] Siclari VA, Qin L. Targeting the osteosarcoma cancer stem cell. J Orthop Surg Res. 2010; 27;5:7810.1186/1749-799X-5-78Search in Google Scholar
[19] Bielack S, Carrle D, Casali PG; ESMO Guidelines Working Group. Osteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20 Suppl 4:137-13910.1093/annonc/mdp154Search in Google Scholar
[20] Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;19;369(9574):1742-175710.1016/S0140-6736(07)60781-8Search in Google Scholar
© 2016 Feng Yun-Bo et al.
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 3.0 License.
