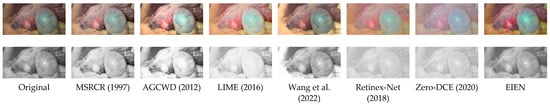
<p>Scale (1–8) for the classification of the different ripening stages of bananas, associated with changes in the color of the peel, following the one proposed by Escalante et al. [<a href="#B25-sensors-24-07010" class="html-bibr">25</a>].</p> Full article ">Figure 2
<p>Experimental configuration implemented for the acquisition of diffuse reflectance spectra for banana tissue and volunteers participating in this study: (<b>a</b>) Mini spectrometer; (<b>b</b>) tungsten halogen light source; (<b>c</b>) personal computer equipment for spectral analysis; (<b>d</b>) bifurcated fiber-optic probe and zoom of the geometries of the optical probes used.</p> Full article ">Figure 3
<p>Shades of skin color ordered in ascending order according to Fitzpatrick’s classification (I–VI). The shades presented are based on those proposed by Caerwyn et al. [<a href="#B37-sensors-24-07010" class="html-bibr">37</a>].</p> Full article ">Figure 4
<p>Areas selected to calculate the average color value of two of the regions of interest measured with DRS. The area covered by the blue box represents the area selected to average the healthy skin color, while the region covered by the red box represents the area chosen to average the color of the nevus. This image corresponds to a volunteer classified with skin phototype II.</p> Full article ">Figure 5
<p>Main layers of the skin. The simulation of the sampling volume for both probes considers two main skin layers. The first layer corresponds to the epidermis, with a thickness of 60 microns, and the second layer represents the dermis, with a thickness of 5000 microns. ‘S’ and ‘D’ stand for “source fiber” and “detector fiber”, respectively. Yellow arrows show the direction of energy emitted from the source fiber into the sample and collected by the detector fiber, while brown arrows illustrate an example of photons’ paths within the tissue.</p> Full article ">Figure 6
<p>Diffuse reflectance spectra of banana fruit skin in two measurement regions with the fiber-optic probe with the largest distance between the centers of the emitting and collecting fibers (homemade probe): (<b>a</b>) Spectral curves of seven ripening stages of an area without a spot of banana fruit skin; (<b>b</b>) spectral response of an area with a spot at different ripening stages.</p> Full article ">Figure 7
<p>Normalized averaged spectral curves obtained from two regions studied on the skin of the banana fruit for samples in seven stages of maturation: (<b>a</b>) Spectra of the skin without the presence of the spots or lesions in a region near to the selected spot, and (<b>b</b>) spectra of the brown spots selected in the same sample.</p> Full article ">Figure 8
<p>Normalized spectral curves recorded with the commercial probe on banana fruit skin spots at four ripening stages (4 to 7).</p> Full article ">Figure 9
<p>Spectral normalized curves obtained in two skin regions of 10 volunteers: (<b>a</b>) Spectra corresponding to a skin region close to the nevus; (<b>b</b>) spectra corresponding to the selected nevi (one per volunteer).</p> Full article ">Figure 10
<p>Normalized spectral curves were obtained from two regions of interest from eight volunteers: (<b>a</b>) Spectra taken in a region of healthy skin in close proximity to the nevus, and (<b>b</b>) spectra corresponding to the nevi measured in each volunteer. In the case of volunteer 8, three measurements were taken, designated as “8-1”, “8-2”, and “8-3”, corresponding to the order in which they were taken.</p> Full article ">Figure 11
<p>(<b>a</b>) Areas of the volunteers’ nevi and brown spots on the skin of the banana fruit. The red circles represent the nevi of the 10 volunteers (volunteer 4 was excluded due to the large area of their nevus, 42.43 mm<sup>2</sup>). Black circles represent the selected spots in the different banana fruit samples. (<b>b</b>) Graph showing the average area of the brown spots according to the ripening or maturation stage of the sample, where the yellow line shows the value of the average area of the nevi (in this calculation the nevus of volunteer 4 was excluded, as indicated before).</p> Full article ">Figure 12
<p>Images of spots on the skin of banana fruit taken from samples of different ripening stages.</p> Full article ">Figure 13
<p>Results of the analysis of the average value of the color intensity of each volunteer ordered by skin phototype according to Fitzpatrick’s classification (see <a href="#sensors-24-07010-f003" class="html-fig">Figure 3</a>), after evaluating two skin regions, from the results of the Silonie Sachveda survey. (<b>a</b>) The first row corresponds to a visually selected area of healthy skin (HS) without the presence of lesions or hair, measured with DRS; (<b>b</b>) region of healthy skin (HS) under similar circumstances in which no DRS measurements were performed (each volunteer is referred by the number 1–10).</p> Full article ">Figure 14
<p>Shades obtained for the volunteers’ nevi as a result of the calculation of the average value of the color intensity in the RGB system.</p> Full article ">Figure 15
<p>Comparison of the average color of nevi, (N1–N10) and spots (Sp1, Sp2, and Sp29) in the banana fruit skin, where the minimum value of ARE between the three relative percentage errors was obtained for each nevus–spot combination. The spots with the greatest similarity were captured in samples with the higher ripening stages: 4, 5, and 6. Furthermore, the actual images of the nevus and the spot are positioned in the top and bottom rows, respectively.</p> Full article ">Figure 16
<p>Spectral comparison between nevi and spots considering the minimum MSE values with the normalized spectra from 400 nm to 750 nm. The first column shows the general spectral comparison, while the second column presents the comparison of the nevus with the spots with the minimum MSE value for the spectral region classified as R (from 571 to 750 nm; range marked by a horizontal red bar in the figures): (<b>a</b>) Spectra of the nevus of volunteer 1 and the Sp23 spot, with an MSE of 0.02, and the Sp22 spot; (<b>b</b>) spectra of the nevus of volunteer 2 and the Sp9 spot, with an MSE of 0.04, and the Sp29 spot; (<b>c</b>) spectra of the nevus of volunteer 4 and the Sp41 spot, with an MSE of 0.04, and the Sp22 spot; (<b>d</b>) spectra of the nevus of volunteer 7 and the Sp23 spot, with an MSE of 0.03, and the Sp21 spot.</p> Full article ">Figure 17
<p>Spectral comparison between nevi and spots measured with the commercial probe, considering the minimum MSE values of the normalized spectra from 400 nm to 750 nm. The first column (<b>a</b>,<b>c</b>,<b>e</b>) presents the three combinations with the maximum MSE values obtained among the minima, while column two (<b>b</b>,<b>d</b>,<b>f</b>) shows the three combinations with the lowest MSE, highlighting mainly (<b>a</b>) nevus of volunteer 7 compared to Sp18 spot, with an MSE of 0.0087, being the maximum MSE value among the minima obtained; and (<b>b</b>) nevus of volunteer 4 compared to Sp22 spot, with an MSE of 0.0010, being the lowest value among those obtained with the commercial probe.</p> Full article ">Figure 18
<p>Cumulative variance explained by different numbers of principal components of nevi and spot spectra. The dashed line indicates the 90% variance threshold, which is obtained from the third component on.</p> Full article ">Figure 19
<p>Spectral comparisons between nevi and spots according to the minimum values of the Mahalanobis distance, calculated from the first five principal components. (<b>a</b>) Comparison between the normalized spectra of the nevus of volunteer 4 and Sp23 spot; (<b>b</b>) comparison between the normalized spectra of volunteer 8’s nevus and Sp24 spot.</p> Full article ">Figure 20
<p>A comparative study of the SV simulation of both probes for light with wavelengths representative of the spectral range of measurement and its impact on the similarity of diffuse reflectance spectra of pigmented lesions of human skin and brown spots on bananas.</p> Full article ">Figure 21
<p>Spectral dependence of the maximum depth of the sampling volume, Z<sup>0</sup><sub>Max</sub>, for the two fiber-optic probes employed in this study using nine discrete wavelengths in the spectral range of interest.</p> Full article ">