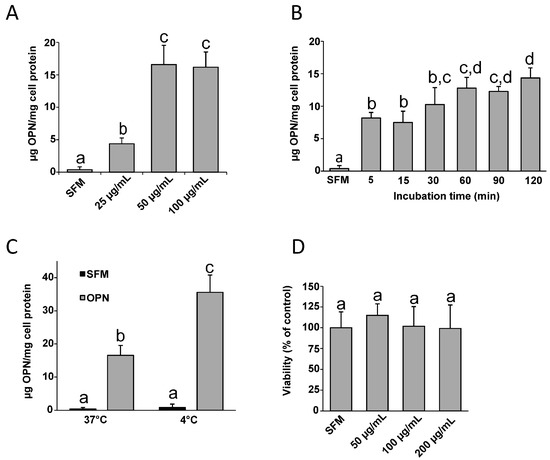
<p>The effect of DOX application on 9 different concentrations obtained by serial dilution in the concentration range of 10-1000 nM in HeLa cervix adenocarcinoma (<b>A</b>–<b>C</b>) and HaCaT human skin keratinocyte cell line (<b>D</b>–<b>F</b>) on compared to the vehicle group and the IC50 value of the chemotherapy agent (n = 6; data are mean ± standard deviation values, inhibition concentration (IC) values calculated by probit analysis). * Data are statistically significant compared to control, one-way ANOVA, Tukey HSD test, <span class="html-italic">p</span> ≤ 0.05.</p> Full article ">Figure 2
<p>Effect of GA application on 9 different concentrations obtained by serial dilution between 10-1000 µM concentration range in HeLa cervix adenocarcinoma (<b>A</b>–<b>C</b>) and HaCaT human skin keratinocyte cell line (<b>D</b>–<b>F</b>) cell lines for 24, 48 and 72 hours on cell viability compared to the vehicle group and the IC50 value of GA (n = 6; data are mean ± standard deviation values, inhibition concentration (IC) values calculated by probit analysis). * Data are statistically significant compared to control, one-way ANOVA, Tukey HSD test, <span class="html-italic">p</span> ≤ 0.05.</p> Full article ">Figure 3
<p>Cell morphology, nuclear structure, and apoptotic body formation (magnification: ×20) in HeLa cervical adenocarcinoma cell populations treated for 48 hours with vehicle control (<b>A</b>,<b>A1</b>), DOX IC50: 137.6 nM (<b>B</b>,<b>B1</b>), GA IC50: 239.2 μM (<b>C</b>,<b>C1</b>), and DOX IC50+GA IC50 (<b>D</b>,<b>D1</b>) (Arrow: apoptotic cell).</p> Full article ">Figure 4
<p>H-scores were derived from semi-quantitative assessments of both staining intensity (scale 0–3) and the percentage of positive cells (0–100%) and, when multiplied, generated a score ranging from 0 to 300.</p> Full article ">Figure 5
<p>Relative fold increases values of P53 and BAX gene expressions in HeLa cervical adenocarcinoma cell lines, DOX IC<sub>50</sub>: 137.6 nM, GA IC<sub>50</sub>: 239.2 μM, 48 h, after single and combined drug administration (data in multiple control with β-actin and GAPDH mRNA level). Method, n = 4 data mean ± SH), * means are statistically different, one-way ANOVA, Tukey HSD test, <span class="html-italic">p</span> values are given in the graph.</p> Full article ">Figure 6
<p>PPI and interaction between various genes of cervical cancer.</p> Full article ">Figure 7
<p>Enrichment analysis for the 530 common compound targets in cancer pathway.</p> Full article ">Figure 8
<p>Enrichment analysis for the 331 common compound targets in human papilloma virüs infection and cervical cancer.</p> Full article ">