Best Practices in Publishing Sampling-event data
Version 2.2
Suggested citation
GBIF (2018) Best Practices in Publishing Sampling-event data, version 2.2. Copenhagen: GBIF Secretariat. https://ipt.gbif.org/manual/en/ipt/3.0/best-practices-sampling-event-data
Introduction
This guide provides details on how to utilize the Darwin Core Archive (DwC-A) format as a means to share sampling-event information in a standard way. It focuses on specific components of the Darwin Core Archive format, and some of the supporting extensions to the core event data class, and provides recommendations on how to best utilize these components to maximize the value of the shared data. This guide does not provide a detailed overview of the Darwin Core Archive format, instead please refer to the Darwin Core Archives How-to Guide.
The DwC-A format and the specific profile described here represent an internationally recognized and ratified data exchange format for sharing sampling-event data. All data exchange standards must strike a balance between the technical scope and capacity on one hand, and social acceptance and uptake on the other. Simple solutions sacrifice coverage and complexity in favour of ease-of-use. Highly complex formats provide more complete solutions for representing any type of data but at the expense of simplicity and require supporting software and expertise. The Darwin Core Archive format represents an intermediate position between the two ends of this spectrum. It focuses on the key elements of sampling-event data and enables an enriched set of data types to be linked to this core structure. The data contained in an archive can be readily understood and used by many ecologists and data managers familiar with basic structured text files. With this international standard, GBIF hopes to facilitate sampling-event data sharing and promote common approaches to cite and recognize the work of the community creating and handling sampling-event data. A standard format also increases relevance and utility.
What is sampling-event data?
Sampling-event data is a type of data available from thousands of environmental, ecological, and natural resource investigations. These can be one-off studies or monitoring programs. Such data are usually quantitative, calibrated, and follow certain protocols so that changes and trends of populations can be detected. This is in contrast to opportunistic observation and collection data, which today form a significant proportion of openly accessible biodiversity data.
How to express sampling-event data in DwC-A?
Darwin Core Archive (DwC-A) is an informatics data standard that makes use of the Darwin Core terms to produce a single, self-contained dataset for checklist data. The collection of files in an archive form a self-contained dataset, which can be provided as a single compressed (Zip or GZIP) file. A dataset is composed of a descriptive metadata document and a set of one or more data files. For more information about DwC-A refer to the Darwin Core Archives How-to Guide.
Sampling-event Metadata
Documenting the provenance and scope of datasets is required in order to publish sampling-event data through the GBIF network. Dataset documentation is referred to as ‘resource metadata’ and enables users to evaluate the fitness-for-use of a dataset. It may describe the sampling methodologies used for its compilation, and the individuals and organizations involved in its creation and management. Metadata is shared in a Darwin Core Archive as an XML document. GBIF provides a metadata profile for sampling-event datasets based on the Ecological Metadata Language. A How-to guide describes all the options for describing a sampling-event dataset using this format. See GBIF Metadata Profile – How-to Guide.
Sampling-event Data
The Darwin Core Archive format provides the structural framework for publishing sampling-event data. Darwin Core Archives consist of a series of one or more text files, in standard comma- or tab-delimited format. The files are logically arranged in a star-like manner with one core file, listing the sampling events (sampling protocol, sample size, location, etc.) surrounded by a number of ‘extensions’, that describe related data types (such as species occurrences, measurements or facts related to the sampling event, etc). Links between core and extension records are made using an event identifier (eventID) data element. In this way, many extension records can exist for each single core event record. This “star-schema” provides a simple relational data model that supports many types of annotations that are common to sampling-event datasets.
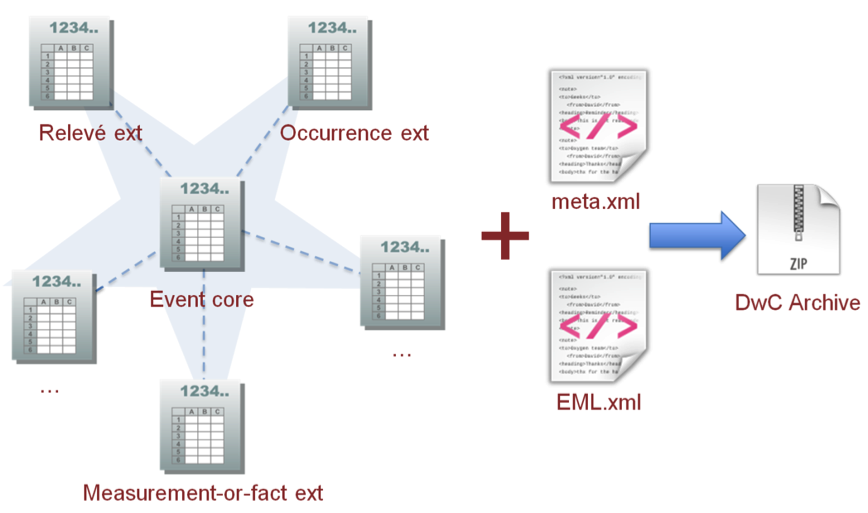
An alternative way to encode sampling-event data is listing the species occurrence in the core file, surrounded by a number of extensions, that describe the related data types (such as measurements relating to the species occurrences, etc). Note listing sampling events in the core file is preferable if a plot or site is the main focus of the study.
Be aware that the current DwC-A star schema such as does have limitations. For example, it does not allow to link measurements and facts to both events and occurrences in the same dataset. Discussion on this prototype extension are taking place on GitHub but there is no solution available yet.
Please check the data quality requirements for sampling events. Note that if you generate your Darwin Core Archive manually, you might need to add a recordID field containing unique identifiers. (This field is generated automatically if you are using an IPT.) For more information on the structure of a Darwin Core Archive, please check the TDWG Darwin Core text guide. You can always check your archive with the GBIF data validator.
Data file formatting recommendations
For ease in understanding, we may use the terms field in this guide to refer to the Darwin Core set of terms in the sampling-event publishing profile to which a users data will be mapped. For example, we will refer to the use of the dwc:scientificName field when referring to the Darwin Core term, scientificName.
-
It is recommended to use TAB or Comma-Separated-Values instead of custom field delimiters and quotes.
-
Be careful and consistent with quotation.
-
Encode text files as UTF-8
-
Make sure you replace all line breaks in a data field, i.e.
\r\nor\r\nwith either simple spaces or use 2 characters like$$to replace\rto escape the line break if the intention is to preserve them. Another option is to replace line breaks with the HTML<br>tag. -
Encode nulls as empty strings, i.e. no characters between 2 delimiters, or
\Nor\NULL, but no other text sequence!
Sample size
The following Darwin Core fields store the sample size of a sampling event:
-
sampleSizeValue: a numeric value for a measurement of the size (time duration, length, area, or volume) of a sample in a sampling event.
-
sampleSizeUnit: the unit of measurement of the size (time duration, length, area, or volume) of a sample in a sampling event.
The value of sampleSizeValue is a number and must have a corresponding sampleSizeUnit. The value of sampleSizeUnit should be restricted to use only SI units/derived units or other non-SI units accepted for use within the SI (e.g. minute, hour, day, litre) as per the Unit of Measurement Vocabulary. Examples are given in Table 1 below.
You may represent a sampling area by using an appropriate WKT shape or latitude/longitude point location. Done correctly, the direction sampling was carried out can also be derived. For example, an ocean trawl line represented using a WKT shape LINESTRING allows the direction of the trawl to be determined based on the standard notation for writing the start and end points.
| sampleSizeValue | sampleSizeUnit |
|---|---|
2 |
hour |
3 |
m2 |
17 |
km |
1 |
litre |
Quantity and abundance
The following Darwin Core fields are also required to be used as a pair:
-
organismQuantity: a numeric or enumeration value for the quantity of organisms.
-
organismQuantityType: the type of quantification system used for the quantity of organisms.
Table 2 lists some example values. The value of organismQuantity is a number or enumeration, e.g., “27” for an organismQuantityType “individuals”, “12.5” for an organismQuantityType “%biomass”, or “r” for an organismQuantityType “BraunBlanquetScale”. The value of organismQuantityType (i.e., the entity being measured) is expected to be drawn from a small controlled vocabulary with terms such as “Individuals”, “%Biomass”, “%Biovolume”, “%Species”, “%Coverage”, “BraunBlanquetScale”, “DominScale”. Examples when combined with organismQuantity values: “+” on DominScale; “5” on BraunBlanquetScale; “45” for %Biomass.
| organismQuantity | organismQuantityType |
|---|---|
14 |
individuals |
r |
BraunBlanquetScale |
0.4 |
%Species |
31 |
%Biomass |
How to uniquely identify sampling events
Each event is uniquely identified using dwc:eventID and occasionally dwc:parentEventID. Although the type and format of identifier is arbitrary we recommend that publishers choose persistent globally unique identifier. In the absence of a GUID, publishers may reuse the original fieldNumber.
Make sure to reuse existing stable identifiers and do not create a new identifier for an event when one is already declared.
How to capture hierarchy of events
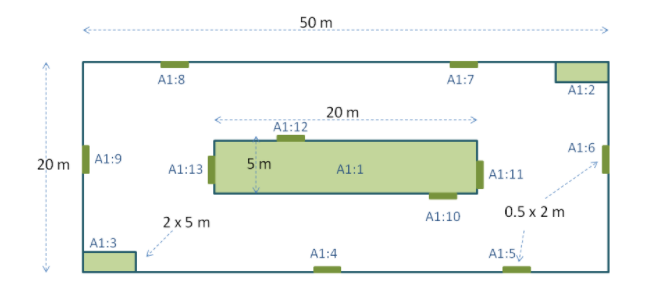
Sampling events can be related to each other (e.g., nested samples) via a common parent identifier. For example, several sub-sampling events within a Whittaker Plot each with their own eventID (e.g., “A1:1”, “A1:2”) would share a common parentEventID (e.g., “A1”) thus enabling them to be linked together easily (see Table 4 and Figure 3).
Further information on the nature of the relationship (e.g. part of a monitoring series) can be described in the project section of the accompanying metadata.
You may also refer to the following FAQ.
How to capture absence data
Refer to the following FAQ.
How to include supplementary multimedia
You may choose to include supplementary media in order to make the data easier to interpret. For example for vegetation data, it is helpful to include a link to the original scanned relevé sheet when interpreting the data.
The files associated have to be hosted on an external server and linked to the occurrence through the dwc:associatedMedia and dwc:associatedReferences. These files may be images, texts or a combination of both as long as the format type is specified. JPG, PNG, etc. images will be visible as thumbnails while PDFs will appear as clickable links.
Publishing sampling-event data
Using GUIDs for identifiers
A number of fields require the usage of unique identifiers: dwc:occurrenceID, dwc:eventID, dwc:organismID and dwc:locationID.
As mentioned previously, although no particular format is enforced, we advise publishers to use Global Unique IDentifiers (GUIDs). There are a few online services which can provide such identifiers. For example, it is possible to use http://www.geonames.org/ to find (or even generate new) identifiers for dwc:locationID, e.g. http://sws.geonames.org/10793757/ is a GUID for a lake in Greenland.
Protect location of sensitive species
If your dataset contains sensitive species, there are several ways to handle it:
-
Simply removing these species from the dataset,
-
Publishing the species identifications at Genus level only,
-
Publishing the sensitive/protected species in a separate dataset,
-
Publish obfuscated sensitive data points in the main dataset and publish non-obfuscated details in an access-limited separate dataset, both datasets including all data records.
Preserving verbatim data
Although verbatim data and descriptions are not visible on the GBIF.org web interface, they are made available to the community through downloads. When entering verbatim description, make sure to link them to the original event or occurrence. For example, the ID or code given to the original event should be entered into dwc:fieldNumber; the ID or code given to the original occurrence observation should be entered into dwc:recordNumber.
Publishing project data as a single dataset
Data produced from a large sampling project should be published as a single dataset if possible. If you must publish multiple datasets, we encourage linking them using a common project identifier in the metadata.
Republishing occurrence data as sampling-event data
Sampling events provide better documentation and benefit both the scientific community and policy makers (read more). We encourage strongly the republishing of occurrence data as sampling-event data when possible.
In order to do so, you should create a new sampling-event dataset and send an email to GBIF’s Help Desk (helpdesk@gbif.org). In this email, you should provide the UUIDs of both the occurrence dataset and the new dataset. We will then be able to link the first dataset to the newest one before de-indexing it thereby avoiding occurrence duplication and preserving citations.
Modelling continuous monitoring of live individuals
If your dataset contains continuous monitoring of live individuals, such as bird tracking data, you can use dwc:organismID to store the ID of the individual being tracked. In addition to that, you should represent each individual being tracked as a single event.
Continuous data quality improvement
Describing sampling-event data in dataset metadata
Publishers should document their dataset as much as possible with a particular emphasis on sampling methodologies.
Besides the mandatory requirements, the metadata should include information about the extent of study, the sampling methods, the quality control and limitations of the study. Although information about fieldwork can be part of the data content, you may describe the sampling location and conditions in the metadata as well.
Examples
Following are some examples of typical sampling-event data sets. In each case, the key fields in the Event core and Occurrence extension are provided. For some examples, additional extensions such as Relevé and measurement-or-fact are also included.
Freshwater invertebrate survey
Core (Event) table
| EventID | samplingProtocol | sampleSizeValue | sampleSizeUnit | eventDate | location | decimalLatitude | decimalLongitude |
|---|---|---|---|---|---|---|---|
C_1428 |
AQEM |
1.25 |
m^2 |
2006-06-21 |
Kinzig O3 Rothenbergen |
50.18689 |
9.100369 |
B_1538 |
AQEM |
1.25 |
m^2 |
2008-11-06 |
Kinzig W3 Bulau |
50.1316 |
8.9657 |
Extension (Occurrence) table
| EventID | scientificName | organismQuantity | organismQuantityType | … |
|---|---|---|---|---|
C_1428 |
Baetis rhodani |
14 |
individuals |
|
C_1428 |
Ephemera danica |
15 |
individuals |
|
C_1428 |
Gyraulus albus |
2 |
individuals |
|
B_1538 |
Serratella ignita |
318 |
individuals |
Explanation
Ephemera danica : A total of 14 individuals from 1.25 square meters were obtained in this sampling event. The derived individuals per sq meter count is 11.2 (14/1.25).
Brackish water invertebrates survey
Core (Event) table
| EventID | samplingProtocol | sampleSizeValue | sampleSizeUnit | startDayOfYear | endDayOfYear | year | location | decimalLatitude | decimalLongitude | … |
|---|---|---|---|---|---|---|---|---|---|---|
IA1 |
hand operated van Veen grab |
0.04 |
m^2 |
147 |
154 |
1995 |
Gialova lagoon |
36.9564 |
21.6661 |
|
IA3 |
hand operated van Veen grab |
0.04 |
m^2 |
147 |
154 |
1995 |
Gialova lagoon |
36.9564 |
21.6661 |
Extension (Occurrence) table
| EventID | scientificName | organismQuantity | organismQuantityType | … |
|---|---|---|---|---|
IA1 |
Abra ovata |
57 |
individuals |
|
IA3 |
Bittium reticulatum |
113 |
individuals |
Extension (Measuremenr-or-Fact) table
| EventID | measurementType | measurementValue | measurementUnit | measurementRemarks | … |
|---|---|---|---|---|---|
IA1 |
Tmp (sed) |
21.5 |
Degree C |
temperature at the bottom surface |
— |
IA1 |
Rdx (sed)0 |
170 |
mv |
Eh value at the bottom surface (0cm) |
— |
Explanation
Abra ovata : A total of 57 individuals from 0.04 square meters were obtained in sampling event IA1.
Each event can also have measurements or facts associated with it, e.g., environmental measurements like sediment temperature and redox potential (Eh).
Macrophyte survey
Note that this example is based on Dutch Vegetation Database (LVD) previous version republished as sampling-event dataset. The Relevé extension underwent significant changes following the publication of the primer. For more information about LVD and the data model for vegetation sampling-event data see: https://gbif.blogspot.com/2016/07/probably-turbovegs-best-kept-secret.html
| EventID | samplingProtocol | sampleSizeValue | sampleSizeUnit | eventDate | location | decimalLatitude | decimalLongitude | … |
|---|---|---|---|---|---|---|---|---|
1001 |
Braun Blanquet |
100 |
m^2 |
09/08/2012 |
Kinzig O3 Rothenbergen |
50.18689 |
9.100369 |
Extension (Occurrence) table
| EventID | scientificName | organismQuantity | organismQuantityType | … |
|---|---|---|---|---|
1001 |
Acer psuedoplatanus |
r |
BraunBlanquetScale |
Extension (Relevé) table
| EventID | syntaxonCode | inclination | coverTotal | treesCover | coverShrubs | highTreeLayerHeight | highHerbLayerMeanHeight | mossesIdentified | … |
|---|---|---|---|---|---|---|---|---|---|
1001 |
843200 |
40 |
100 |
95 |
50 |
25 |
40 |
Y |
— |
Explanation
Acer psuedoplatanus : In the 100 sq meters surveyed, the abundance of the species was reported as “r” on the Braun Blanquet scale.
Additional vegetation plot measurements such as vegetation community type (syntaxon) % coverage values that are typical of TurboVeg type databases are captured in a Relevé (vegetation-plot) extension.
Lepidoptera survey I
Core (Event) table
| EventID | samplingProtocol | sampleSizeValue | sampleSizeUnit | startDayOfYear | endDayOfYear | year | location | decimalLatitude | decimalLongitude | … |
|---|---|---|---|---|---|---|---|---|---|---|
2320 |
Jalas-model light trap with 160W ML matt lamp |
16 |
day |
164 |
180 |
1999 |
Kungsmarken |
55.72 |
13.28 |
… |
Extension (Occurrence) table
| EventID | scientificName | organismQuantity | organismQuantityType | … |
|---|---|---|---|---|
2320 |
Opisthograptis luteolata |
11 |
individuals |
Explanation
Opisthograptis luteolata : 11 individuals were observed over the sampling period of 16 days. The derived number of individuals per day is 0.68 (11/16).
Lepidoptera survey II
Core (Event) table
| EventID | samplingProtocol | sampleSizeValue | sampleSizeUnit | eventDate | location | decimalLatitude | decimalLongitude | … |
|---|---|---|---|---|---|---|---|---|
1014-tr023m |
Pollard walks |
250 |
m^2 |
2012-10-11 |
Ramat Hanadiv botanik garden |
32.553191 |
34.947492 |
|
1012-tr006-s5 |
Pollard walks |
250 |
m^2 |
2012-05-02 |
Carmel Hurshan haarbaim |
32.75789805 |
35.02697333 |
Extension (Occurrence) table
| EventID | scientificName | organismQuantity | organismQuantityType | … |
|---|---|---|---|---|
1014-tr023m |
Pieris rapae |
1 |
individuals |
|
1014-tr023-s5 |
Maniola telmessia |
2 |
individuals |
Extension (Measurement-or-Fact) table
| EventID | measurementType | measurementValue | measurementUnit | measurementRemarks | … |
|---|---|---|---|---|---|
1014-tr023m |
Temp |
20 |
Degree C |
||
1014-tr023m |
Wind speed |
light |
|||
1014-tr023m |
Cloudiness |
0 |
Level 1 of 8 |
||
1014-tr023m |
AvgAltitude |
10 |
m |
Average altitude |
Explanation
Pieras rapae : A total of 1 individual from 250 sq metres was obtained in this sampling event. Several environmental measurements (e.g., temperature, wind speed, cloudiness) are included in a measurement-or-facts extension.
Reef fish survey
Core (Event) table
| EventID | samplingProtocol | sampleSizeValue | sampleSizeUnit | eventDate | location | decimalLatitude | decimalLongitude | … |
|---|---|---|---|---|---|---|---|---|
506003329 |
Reef Life Survey methods |
500 |
m^2 |
2006-09-02 |
Cocos Islands |
5.56187 |
-87.04693 |
|
57003326 |
Reef Life Survey methods |
500 |
m^2 |
2006-12-11 |
Panama Bight |
4.008553 |
-81.605377 |
Extension (Occurrence) table
| EventID | scientificName | organismQuantity | organismQuantityType | … |
|---|---|---|---|---|
506003329 |
Acanthurus nigricans |
42 |
individuals |
|
506003329 |
Acanthurus xanthopterus |
1 |
individuals |
|
506003329 |
Aulostomus chinensis |
4 |
individuals |
|
506003329 |
Axoclinus cocoensis |
1 |
individuals |
Explanation
Aulostomus chinensis : A total of 4 individuals from 500 sq metres were obtained in this sampling event.
Nested samples
| EventID | parentEventID | samplingProtocol | sampleSizeValue | sampleSizeUnit | eventDate | location | decimalLatitude | decimalLongitude | … |
|---|---|---|---|---|---|---|---|---|---|
A1 |
Modified Whittaker Plot |
1000 |
m^2 |
1984-03-18 |
Monarch |
55.72 |
13.28 |
||
A1.1 |
A1 |
100 |
m^2 |
||||||
A1.2 |
A1 |
10 |
m^2 |
||||||
A1.3 |
A1 |
10 |
m^2 |
||||||
A1.4 |
A1 |
1 |
m^2 |
||||||
A1.5 |
A1 |
1 |
m^2 |
||||||
A1.6 |
A1 |
1 |
m^2 |
||||||
A1.7 |
A1 |
1 |
m^2 |
||||||
A1.8 |
A1 |
1 |
m^2 |
||||||
A1.9 |
A1 |
1 |
m^2 |
||||||
A1.10 |
A1 |
1 |
m^2 |
||||||
A1.11 |
A1 |
1 |
m^2 |
||||||
A1.12 |
A1 |
1 |
m^2 |
||||||
A1.13 |
A1 |
1 |
m^2 |
Additional information that could also be included or was previously included
The Event core elements are mainly drawn from the DwC classes Event, Location and Geological Context (Table 3). The Occurrence extension elements are drawn from the Occurrence, Taxon and Identification classes. For reasons of consistency, the Occurrence extension includes all terms found in the Occurrence core. Thus Event, Location and Geological Context terms are also listed for the Occurrence extension but are actually redundant. Note the IPT hides redundant terms by default when mapping for the user’s convenience.
Event Core |
eventID, parentEventID, samplingProtocol, sampleSizeValue, sampleSizeUnit, samplingTaxaRange, siteTreatment, siteID, layer |
|---|---|
Occurrence Extension |
eventID, organismQuantity, organismQuantityType, siteID+, layer+ |
The "+" symbol indicates proposed new terms not yet ratified.