A slurm based schema for RNA-seq analysis to execute on Linux clusters. A Pipeline:4-RNA-seq and a minimal RNA-seq cook book to explain each step is freely available here. This book was based on the old Taito server. For practical use read the below instructions.
The purpose of this project is to develop an easily customizable commandline based schema "for RNAseq" analysis. Additionally, it has basic Linux scripts for file manipulation which is a bottleneck for developing a new command line pipeline.
An alternative of 4-RNA-seq is nf-co.re/rnaseq. However, it can not perform pseudo alignment where reference genome is missing. Therefore it is still relevant.
- SortmeRNA:
- Now possible to give multiple fasta files as reference.
- Tuned for disk utilization and no need for a separate command to compress fastq files.
- pipeline.sh: single command to finish the whole pipeline.
- errors handling: halt and send the right job status by email after an error in any of the tasks from sbatch_commandlist.
- version.txt: tracks the versions of the used software for future reference.
4-RNA-seq is required to download every new project.
Download
For each experiment, the 4-RNA-seq pipeline needs to be downloaded separately. Let us download it to a directory named "myProjectDir" with the following commands
git clone https://github.com/vondoRishi/4-RNA-seq myProjectDirFrom now on myProjectDir is the project space
Prepare the workspace
Make a directory "rawreads" inside "myProjectDir" and copy fastq(.gz) files there.
mkdir myProjectDir/rawReads
cp sourceDir/*fastq.gz myProjectDir/rawReadsThis pipeline and workflow is optimized for Puhti server. The old pipeline for Taito.csc server is moved at "Taito" branch due to decommisioning of the old server.
Please read the server documentation before start using. It may need to customized according to that.
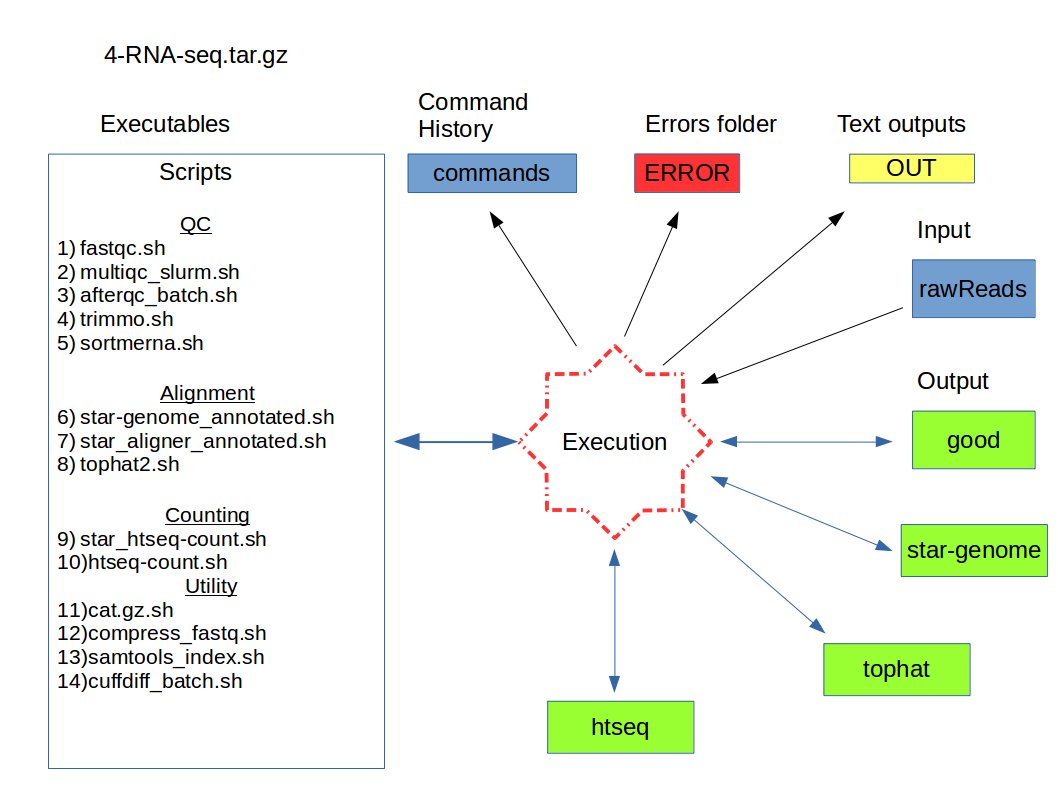
The objective of this documentation is to make execution faster and reproducible as much as possible. The project folder ( should be in /scratch/<project> path and in this example "myProjectDir" ) should contain these folders before starting
- scripts : contains all scripts to run in taito server
- OUT : contains output files from all scripts
- ERROR : contains error files from all scripts
- commands : contains actual commands { will be required in future to find the project specific parameters }
- rawReads : should contain sequencing reads generated by the sequencing machine. Folder name could be anything.
Additional info : Library type, sequencing platform Input: Reference Genome (DNA sequences) fasta and annotation file (GTF) Run “ls -lrth” after every step to find the last modified file
Need to install afterqc by the users of puhti.csc.fi.
Before execution please define the project variables inside 4-rna-seq.config file.
These values will be used by different scripts of this pipeline.
cd myProjectDirBefore execution please confirm that there are enough space
If the varibles are defined correctly then with the single command QC and transcript/gene quantification can be performed.
bash pipeline.shThis will execute FastQC, AfterQC, Sortmerna, Salmon and MultiQC. After Completion of each step it will send an email.
Do not forget to check any error/failure message from any of the programs
grep -rnwi ERROR OUT -e 'error\|fail\|Disk.*'- Start QC ( quality checking) with Fastqc and Multiqc. The scripts/fastqc.sh executes first Fastqc and then Multiqc internally.
Input : directory rawReads with fastq or fastq.gz files
Execution: Replace <project> and <email_id>.
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/fastqc.sh rawReads # Don't use "rawReads/" Output : directory rawReads/rawReads.html and other files
ls -lrth rawReads/- Filter/trimminging with
a) AfterQC (Define the "AfterQC" variable in 4-rna-seq.config before using)
Input : directory rawReads with fastq or fastq.gz files
Execution :
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/afterqc_batch.sh rawReads Output : directory good, bad and QC
b) ALERT for "single end reads" users!! AfterQC can not trim adapters from single end reads. Hence use Trimmomatic to cut adapters [ check for trimming parameters ] [ Tips for filename ]
Input : directory good with fastq or fastq.gz files
Execution:
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/trimmo.sh good trimmed_reads Output : directory trimmed_reads
{ Run step 1 review effect of trimming }
-
Sortmerna.sh [ We can also execute this at the very beginning (optional) ]
Sometimes ribosomal or any other unwanted RNAs may present in the library. Sortmerna could be used to filterout them.
Confirm the parameters in file 4-rna-seq.config- "sortMeRNA_ref" path to a folder to reference rRNA fasta files.
First index the reference fasta file
If the indexing is available then the program will do nothing.
Input: sortMeRNA_ref variable in 4-rna-seq.config
Execution:sbatch -A <project> -D $PWD --mail-user <email_id> scripts/sortMeRNA_indexdb.sh
Now filter out
Input: good
Execution:sbatch -A <project> -D $PWD --mail-user <email_id> scripts/sortmerna.sh good sortMeRna
Output: sortMeRna, the folder contains many different types of file. Fastq/fq files starting with non_Rna will be used in downstream analysis. Files with .log will be used by multiqc to summarize. The "rRna" fastq/fq and ".sam" files are removed by default from sortMeRna before next step. To retailn these files comment out "rm -rf $2/rRna_*{fastq,fq}"
Finally summarize the presence of rRNA.
Execution:sbatch -A <project> -D $PWD --mail-user <email_id> scripts/fastqc.sh sortMeRna
Output: sortMeRna
-
Salmon
Confirm the parameters in file 4-rna-seq.config- "transcripts" path to reference transcripts
- "gene_annotation" path to gtf file
Input: sortMeRna (any directory containing fastq files) Execution:
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/salmon.sh sortMeRna salmon_index salmon_quant
Output: salmon_index = transcriptome indices
salmon_quant = provides a sub-directory with quant.sf for each input samples
Till now we have generated multiqc reports for every command or folder. Now to summarize all in one place execute. Edit multiqc configuration file if requires
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/multiqc_slurm.shsalmon_quant is the final out put folder.
Archieve the important folders
It is an example command only.
tar -czvf results_export.tar.gz *.html *.gtf* *.fa* salmon_quant/ 4-rna-seq.config version.txtDepending upon the library preparation kit the parameters of alignment software need to set. Here are few examples of different popular library kits. [ please report any missing library type and parameters]
To align to a reference genome
-
STAR:
Confirm the parameters in file 4-rna-seq.config- "maxReadLength" to maximum read length
- "genome_file" to path to reference genome
- "gene_annotation" path to gtf file
Input: folder which contains the filtered reads; ex. good or sortMeRna
Execution:sbatch -A <project> -D $PWD --mail-user <email_id> scripts/star.sh good star_alignment
Output: star_alignment (contains bam files and quality report star_alignment.html)
-
htseq-count
[ STAR can also give count values of htseq-count’s default parameter but htseq-count will be used separately]
Confirm the parameters in file 4-rna-seq.config- "stranded" depending upon the library type
- "gene_annotation" path to gtf file
Input: star_alignment
Execution:sbatch -A <project> -D $PWD --mail-user <email_id> scripts/star_htseq-count.sh star_alignment star_count
Output: count values at star_count/htseq_*txt and quality report at star_count.html
Need to sort (uncomment for tophat output bams) and index.
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/samtools_index.sh bam_directoryThere are, some times, multiple copies of same sample from multiple runs. It will be easier to concatenate all these multiple copies in single fastq.gz files before starting any workflow or 4-RNA-seq. Let us assume there are two samples control_1 and treated_1 and they have two copies from two separate runs, run_1 and run_2. Therefore, in the project directory there should have two sub-directories
-
run_1
- control_1_run2019.fastq.gz
- treated_1_run2019.fastq.gz
-
run_2
- control_1_run2020.fastq.gz
- treated_1_run2020.fastq.gz
Additionally another file, such as, sample_names.txt containing each sample names in a separate line. In this case the sample file should look like this
control_1
treated_1
Now we can use
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/cat.gz.sh sample_names.txt rawReadscat.gz.sh will search any fastq.gz files matching with names given in sample_names.txt in ALL sub-directories and concatenate them. The output files can be found in rawReads directory.
sbatch -A <project> -D $PWD --mail-user <email_id> scripts/compress_fastq.sh old_datasbatch scripts/cuffdiff_batch.sh Derm Ctrl Fgf20 star-genome_annotated