This repository has been moved to GitHub.
We provide a deep-learning method to better estimate the size of occluded objects. The method is based on ORCNN (https://github.com/waiyulam/ORCNN), which is an extended Mask R-CNN network that outputs two masks for each object:
- The regular visible mask (purple mask below)
- An additional amodal mask of the visible and the occluded pixels (green mask below)

See INSTALL.md
The deep-learning method that can be used to estimate the diameter of occluded crops:
ORCNN.md
The "base-line" method, which is based on Mask R-CNN and a circle fit method. This method can be compared to the ORCNN sizing method:
MRCNN.md
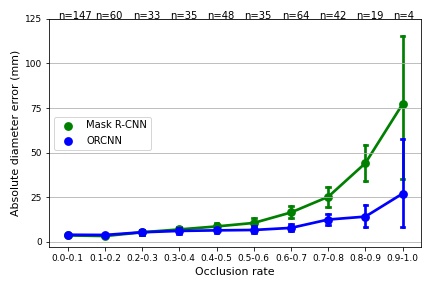
We evaluated the sizing performance of the two methods on an independent test set of 487 RGB-D images. The broccoli heads in the test set had occlusion rates between 0% and 100%.
The table and the graph below summarizes the average absolute diameter error (mm) for 10 occlusion rates. The number between the brackets is the standard deviation (mm).
| Occlusion rate | Mask R-CNN | ORCNN | P-value Wilcoxon test |
|---|---|---|---|
| 0.0 - 0.1 (n=147) | 3.6 (3.1) | 4.0 (2.9) | 0.10 (ns) |
| 0.1 - 0.2 (n=60) | 3.2 (2.6) | 3.9 (2.4) | 0.06 (ns) |
| 0.2 - 0.3 (n=33) | 5.3 (4.1) | 5.4 (4.0) | 0.64 (ns) |
| 0.3 - 0.4 (n=35) | 7.0 (4.8) | 6.1 (4.5) | 0.39 (ns) |
| 0.4 - 0.5 (n=48) | 8.6 (7.0) | 6.4 (4.7) | 0.09 (ns) |
| 0.5 - 0.6 (n=35) | 10.6 (7.8) | 6.6 (6.0) | 0.02 (*) |
| 0.6 - 0.7 (n=64) | 16.5 (13.6) | 7.8 (7.8) | 0.00 (****) |
| 0.7 - 0.8 (n=42) | 25.2 (18.4) | 12.5 (10.2) | 0.00 (***) |
| 0.8 - 0.9 (n=19) | 44.1 (24.0) | 14.1 (13.7) | 0.00 (***) |
| 0.9 - 1.0 (n=4) | 77.2 (43.2) | 27.0 (27.5) | - |
| All (n=487) | 10.7 (15.3) | 6.5 (7.3) | 0.00 (****) |
- : too few samples, ns : P> 0.05, * : 0.01 < P <= 0.05, ** : 0.01 < P <= 0.05, *** : 0.001 < P <= 0.01, **** : P <= 0.001
We have made our image-dataset publicly available under the NonCommercial-ShareAlike 4.0 license (CC BY-NC-SA 4.0). This means that our dataset can only be downloaded and used for non-commercial purposes. Please check whether you or your organization can use our dataset: https://creativecommons.org/licenses/by-nc-sa/4.0/
Our dataset consists of 1613 RGB-D images, including annotations and ground-truth measurements: https://doi.org/10.4121/13603787
| Network | Backbone | Dataset | Weights |
|---|---|---|---|
| Mask R-CNN | ResNext_101_32x8d_FPN_3x | Broccoli | model_0008999.pth |
| ORCNN | ResNext_101_32x8d_FPN_3x | Broccoli | model_0007999.pth |
Our software was forked from ORCNN, which was forked from Detectron2. As such, our CNN's will be released under the Apache 2.0 license.
Please cite our research article or dataset when using our software and/or dataset:
@article{BLOK2021213,
title = {Image-based size estimation of broccoli heads under varying degrees of occlusion},
author = {Pieter M. Blok and Eldert J. van Henten and Frits K. van Evert and Gert Kootstra},
journal = {Biosystems Engineering},
volume = {208},
pages = {213-233},
year = {2021},
issn = {1537-5110},
doi = {https://doi.org/10.1016/j.biosystemseng.2021.06.001},
url = {https://www.sciencedirect.com/science/article/pii/S1537511021001203},
}
@misc{BLOK2021,
title = {Data underlying the publication: Image-based size estimation of broccoli heads under varying degrees of occlusion},
author = {Pieter M. Blok and Eldert J. van Henten and Frits K. van Evert and Gert Kootstra},
year = {2021},
publisher = {4TU.ResearchData},
doi = {https://doi.org/10.4121/13603787},
url = {https://data.4tu.nl/articles/dataset/Data_underlying_the_publication_Image-based_size_estimation_of_broccoli_heads_under_varying_degrees_of_occlusion/13603787},
}
The size estimation methods were developed by Pieter Blok.