Note: The latest versions of the model have been fit to age-structured data, part of which are not publicly available. We have added recent versions of the model here with updated code. However, to ensure HIPAA compliance, we are not adding the data files to this repository. For the most recent projections, see the Santa Cruz County dashboard.
The Santa Cruz County (SCZ) COVID-19 model is a time-discrete, stochastic SEIR model that uses Bayesian statistical methods, such as Hamiltonian Markov Chain Monte Carlo (MCMC) simulations, to forecast the COVID-19 pandemic in Santa Cruz County, California. The model requires a set of parameters, equations, and local data to help inform its simulations. The model is set to run 4,000 simulations and fine-tune the inputted parameters using the local data (confirmed COVID-19 hospitalizations, confirmed COVID-19 cases, and deaths). The model projects a range of different scenarios that fit the inputs provided and are displayed in the exported plots.
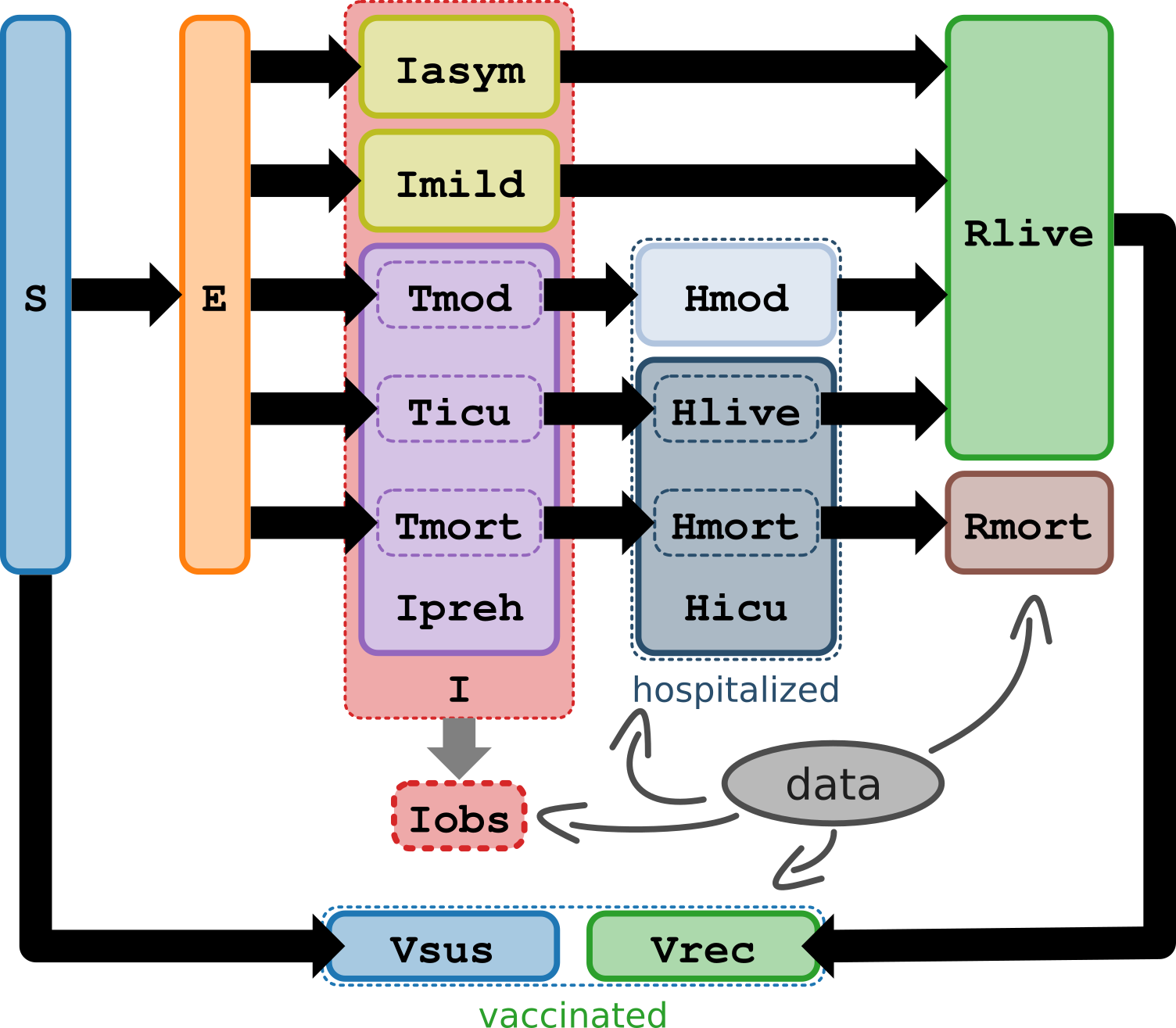
The model contains 11 compartments to divide COVID-19 cases into the asymptomatic, mild, and moderate to severe illness, which better informs hospitalization and death projections (see diagram below).
The Jupyter template notebook can be found here.
Note: If you have issues, questions or find a bug please create an issue in GitHub (above).
- The current model is started on May 1 2021, with initial conditions estimated from a previous model simulation.
- The model's contact rate adjusts every 5 days using spline interpolation.
- A fraction of vaccinated individuals gain immunity.
- COVID-19 cases who recover gain short-term immunity.
- COVID-19 cases can be infectious 2 to 3 days prior to symptom onset.
- COVID-19 hospitalization, ICU, and death rates are calculated based on the overall age demographics of Santa Cruz County.
- Not everyone who tests positive for COVID-19 goes to the hospital.
- COVID-19 cases only die within the hospital.
- Hospitalized COVID-19 patients have a shorter duration of their infectious period because they are less likely to expose others. However, they likely will shed live virus longer, especially if immuno-compromised.
- The model does not account for spatial or network patterns.
- Updating this repository with the latest version of the SCZ COVID-19 Model, the previous version has been moved to v9.
- The model is now using the earliest known date of infection for the case data (also known as episode date). Due to laboratory reporting delays and the transition to episode date, the latest entries in a case count file are likely to be underestimates. Existing daily case counts from the week prior are often updated with new data each day. For the time being, the last entries are not added to the case count file when running the Santa Cruz County model to minimize the impact of this bias.
- In preparation for switching from case counts based on the date lab result were received, to the earliest known date of infection (now reported on the Santa Cruz County dashboard) to fit the model, case count and death data are no longer required to use the same dates and can be specified in separate files.
- Improved data input allows more flexible specification of how to read data from csv or Excel data files. Non-csv text files are no longer supported.
- Removed temporary disclaimer from 2020-08-09.
- Added temporary disclaimer to the plots stating that due to a problem with the State of California’s CalREDIE reporting system, cases have been underreported and projected results likely represent underestimates.
- This problem is affecting the data being fed into the recent projections, not the model code.
- Priors for model misfit parameters (
lambda_Iobs,lambda_Hmod,lambda_Hicu,lambda_Rmort) can now be set by the user. - Santa Cruz and default parametrization now use a tighter fit to mortality data and a looser fit to case count.
- Mean of the prior for rate of mortality has been changed from 1% to 0.5% based on observed mortality in Santa Cruz County.
The core of the model is written in Stan and a Jupyter notebook is used for reading in data, running the model and visualizing its output. Installation instructions are provided here.
In order to provide a new dataset or change the model parameters, edit the beginning of the notebook where each input parameter and the required data files are described in detail.
All parameters listed below can be adjusted by the user. The values below are used by older versions of the model (v9) and by the model that generated the initial conditions for the current model version; the latter now uses the posterior parameter estimates generated by the former.
| Parameter | Value and Distribution | Literature |
|---|---|---|
| Initial Contact Rate | normal(0.2, 0.02)[1] | N/A |
| Latent Period | normal(5, 2) | Cascella et al; Li et al |
| Asymptomatic Infections Days to Recover | normal(7, 5) | He et al |
| Mild Infections Days to Recover | normal(7,4) | He et al |
| Days to Hospital | normal(5,1) | Ferguson et al |
| Non-ICU Cases Days in Hospital | normal(7,1) | Ferguson et al |
| ICU Cases Days in Hospital | normal(16, 1) | Ferguson et al |
| Fraction Tested | time-dependent[2] | N/A |
| Fraction of moderate cases (Hospitalized but non-ICU) | Dirichlet with mean 0.07[3] | Stanford model; Ferguson et al; Verity et al |
| Fraction of severe cases (ICU and alive) | Dirichlet with mean 0.02[3] | Stanford model; Ferguson et al; Verity et al |
| Fraction of severe cases (ICU and dead) | Dirichlet with mean 0.005[3] | N/A |
| Fraction Asymptomatic cases | Dirichlet with mean 0.178[3] | Nishiura et al; Mizumoto et al |
| Fraction of mild cases (non-hospitalized) | Dirichlet with mean 1-sum(0.07, 0.02, 0.005, 0.178)[3] | N/A |
| Population of Santa Cruz County | 273, 213 | https://www.census.gov/quickfacts/santacruzcountycalifornia |
[1] A time-dependent contact rate is then estimated from the data using an AR(1) process and spline interpolation.
[2] Currently increased over time using key dates, see old notebook for details.
[3] All 5 fractions are drawn from the same Dirichlet distribution.
This project is licensed under the MIT License - see the LICENSE file for details.