bigclust is a set of tools for interactively exploring clusterings via dendrograms with several 100k leafs.
For that we are making use of the pygfx WGPU-based rendering engine.
bigclust_demo3.mov
For now the recommended way of installing this package is this:
- Clone the repository:
git clone https://github.com/flyconnectome/bigclust.git
- Install in "editable" mode:
cd bigclust pip install -e .
With this setup, you can just git pull to update the package.
I imagine the typical usage will be to run a big one-off clustering on a remote cluster node and then
load that clustering into bigclust on a local machine. Therefore, bigclust is designed to work with data artefacts
rather than running the clustering itself.
Minimally, you will need:
- A linkage describing the dendrogram.
- A pandas DataFrame with meta data (labels, cluster assigment, etc.) for the original observations.
We are using bigclust for clustering large number of neurons. Therefore bigclust also provides a Neuroglancer-like widget to explore
morphology and a connectivity widget.
Let's illustrate the usage with a simple toy example using the cocoa package for comparative connectomic analyses:
import cocoa as cc
import numpy as np
# Co-cluster two cell types in FlyWire left vs right
cl = cc.generate_clustering(fw=['DA1_lPN', 'DA2_lPN']).compile()# Get the linkage (this is a simple scipy linkage)
Z = cl.get_linkage(method='ward')
np.save("linkage.npy", Z, allow_pickle=False)
# Prepare a table with details we can use as e.g. labels in the dendrogram
t = cl.to_table(cl.extract_homogeneous_clusters(max_dist=2, min_dist=.1, linkage=Z), linkage=Z)
# Save and make sure the order is the same as in our distance matrix
t.set_index("id").reindex(cl.dists_.index).reset_index(drop=False).to_feather("cosine_table.feather")Feel free to use more sensible file names. If you do, you have to adjust the code below accordingly.
Open a new python script - name it e.g. run_bigclust.py:
import pandas as pd
import numpy as np
import trimesh as tm
import bigclust as bc
from wgpu.gui.auto import run
from bigclust._neuroglancer import NglViewer
if __name__ == "__main__":
print("Loading data...", flush=True, end="")
# Load the linkage matrix
Z = np.load("linkage.npy")
# This is the table with the neuron information, including the clusters
table = pd.read_feather("cosine_table.feather")
# Add source information -> we need this to load the neuron meshes in the Neuroglancer viewer
table["source"] = "precomputed://gs://flywire_v141_m783"
# Here we define the default colors for the neurons
table["color"] = table.dataset.map(
{"FwR": "cyan", "FWL": "lightskyblue"}
)
print(" Done.", flush=True)
print("Making dendrogram...", flush=True, end="")
# Now we will instantiate the dendrogram
fig = bc.Dendrogram(
Z,
table=table,
labels='label',
leaf_types='dataset',
clusters='cluster',
ids='id',
)
fig.show()
# Some tweaks:
fig.size = (fig.canvas.screen().size().width(), 300) # make the dendrogram fill the width of the screen
fig.canvas.move(0, 0) # nove it into the top left corner
fig.font_size = 6 # slight larger font size
fig.label_vis_limit = 300 # show more labels before hiding all
fig.leaf_size = 3 # slightly larger leaf size
fig.set_yscale(100) # make the dendrogram a bit taller
print(".", flush=True, end="")
# Load the neuropil mesh for FlyWire from Github
# We will add this to the viewer to make navigation easier
# You could download and store it locally if you want to
neuropil_mesh = tm.load_remote(
"https://github.com/navis-org/navis-flybrains/raw/main/flybrains/meshes/FLYWIRE.ply"
)
# Instantiate the viewer
ngl = NglViewer(table, neuropil_mesh=neuropil_mesh)
ngl.viewer.size = (ngl.viewer.canvas.screen().size().width(), 500)
ngl.viewer.canvas.move(0, 400)
# Tell the dendrogram to sync with the viewer
fig.sync_viewer(ngl)
print(" Done!", flush=True)
# Run the app
# Note: this is only necessary if we're running bigclust from a script
run()Make sure to adjust the filepaths if necessary.
Make sure you have all the data artifacts (linkage.npy and cosine_table.feather) in the same folder as
the run_bigclust.py script. Then:
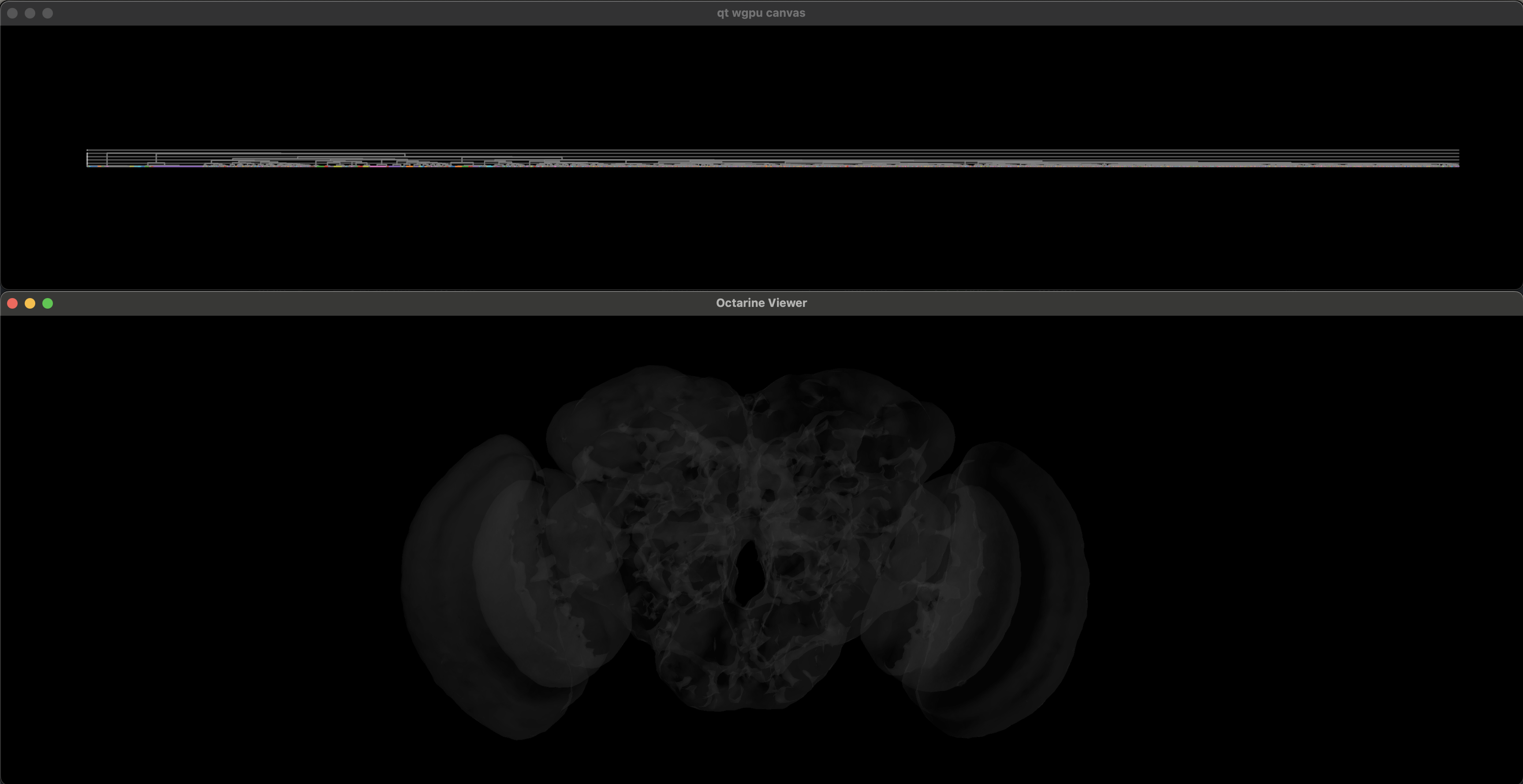
python run_bigclust.pyYou should now be seeing something like this:
The top window contains the dendrogram, the bottom is your 3D viewer.
Dendrogram controls:
- scroll up/down to zoom in/out
- mouse drag to move around
- shift+drag to select neurons (they should appear in the viewer)
escapeor double-click to deselect- hover over a dendrogram leaf to show extra information (see
hover_infoparameter)
3D viewer controls:
- scroll up/down to zoom in/out
- mouse drag to rotate
- two-finger (or middle mouse button) mouse drag to pan
1/2/3to center the view to frontal/dorsal/lateral
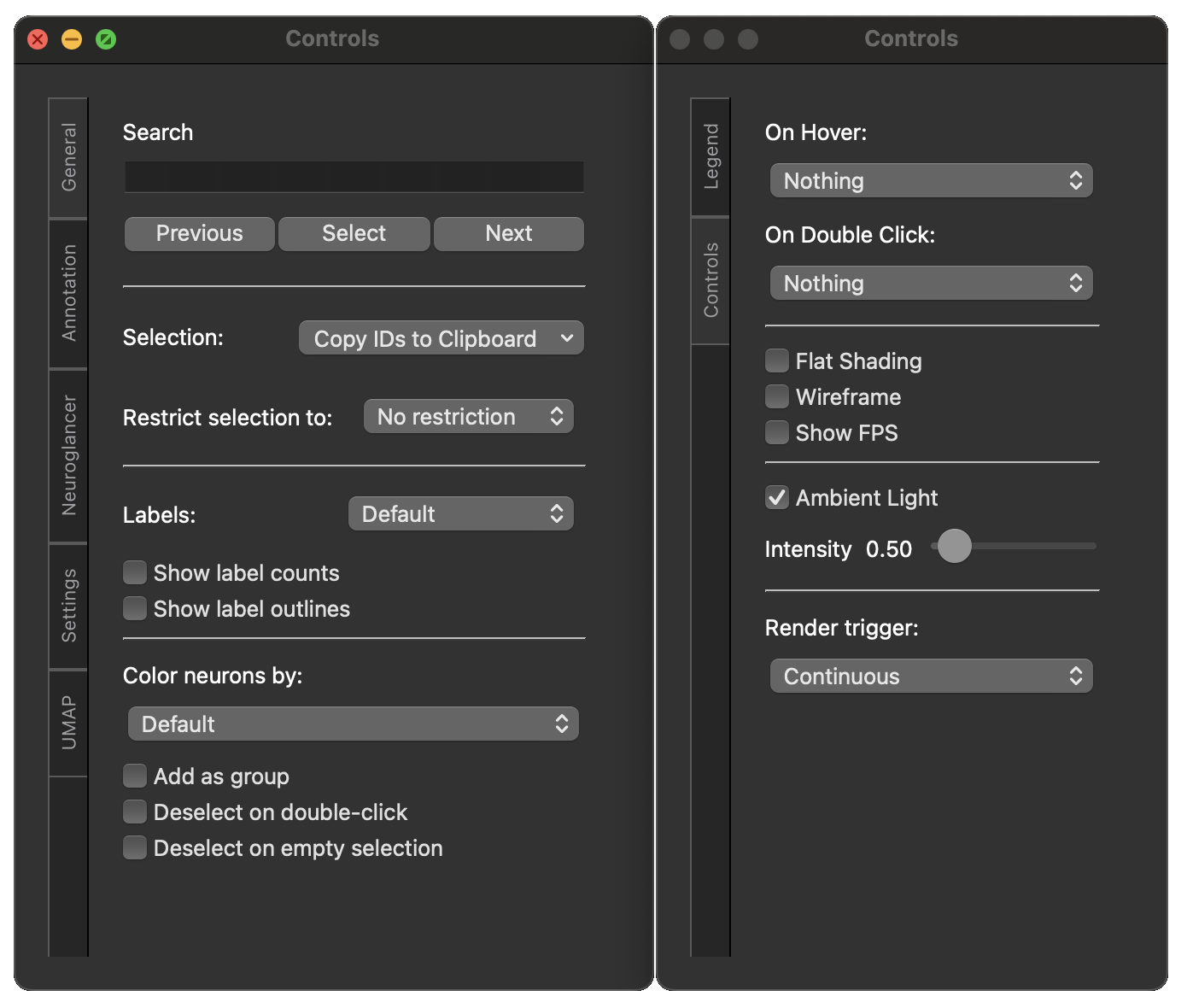
In addition to the above, you can press C while either the viewer or the dendrogram window
is active to bring up a GUI control panel.