The main goal of this package is to make your life easier if you want to:
- Inspect a dataset and compute metrics about its columns content (auto type inference: numeric, mono-label or multi-label).
- Filter the dataset one some criteria (minimum label occurrence, empty example).
- Balance the dataset (TODO) in order to get better performance while training ML or NN models.
- Python 2.7 or 3.6
- Numpy and Pandas
- Command line inspection
You can gain insight into your dataset, getting, among other things, label count, max/min/mean label occurrences (for mono and multi label columns).
# Dataframe with numeric, mono-label, or multi-label (list, tuple, set) columns
df = pd.DataFrame(...)
# Filter label not occurring much in column 'B'
from datadez.filter import filter_small_occurrence
df = datadez.filter.filter_small_occurrence(df, column_name='B', min_occurrence=3)
# Filter empty row based on column 'B' or 'C' values
from datadez.filter import filter_empty
df = filter_empty(df, column_names=['B', 'C'])
# Compute some metrics about your dataset
import pprint
from datadez.summarize import summarize
df_summaries = summarize(df)
pprint.pprint(df_summaries)- Visual inspection
# Dataframe with a multi-label column 'C'
df = pd.DataFrame(...)
# Compute a plotly figure from this dataframe
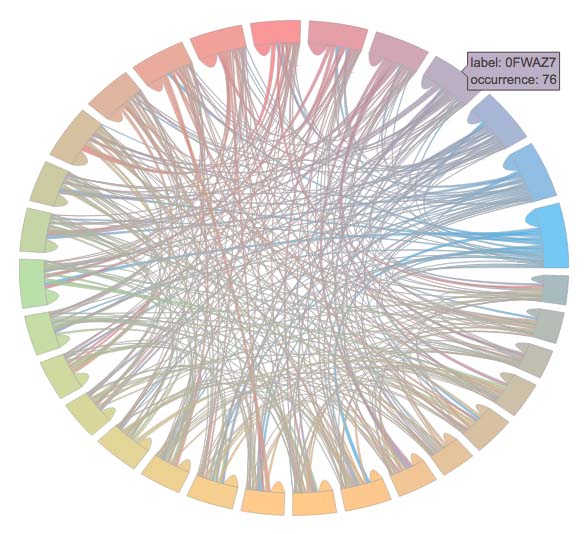
from datadez.dataviz import multilabel_plot
figure = multilabel_plot.intersection_matrix(df, 'C')
# Plot the figure in a file. You can also do this inside a Jupyter notebook
from plotly.offline import plot
plot(figure, filename='chord-diagram.html')Output will look like this:
- Transformation
With this code snippet:
# Dataframe with numeric, text, mono-label, multi-label (list, tuple, set) columns
df = pd.DataFrame(...)
print("Original dataset:")
print(df.head())
# Vectorize text, mono-label and multi-label columns
from datadez.transform import vectorize_dataset
df, vectorizers = vectorize_dataset(df)
print("Vectorized dataset:")
print(df.head())One will get:
Original dataset:
A B C D
0 -0.585248 W2SIF2 [] house jumps adorable crazily
1 0.569125 RYKAXC [IRX7HF, AXQU0L, PM1E1Q, 1FCWZQ] car swims odd merrily
2 -0.076040 7UVFIJ [60WILH, NT28YD, 8IYE5F, 7UVFIJ] monkey barfs clueless dutifully
3 -0.098878 U9WN5M [KS5EXD, YGTPR9] boy runs odd dutifully
4 0.952773 SK1Z1M [AXQU0L] boy barfs odd crazily
Vectorized dataset:
A C ... D
value 1BNK1S 1FCWZQ 246K1M 3A48BH 60WILH 6C3VOQ 6LGS3T 7UVFIJ 8IYE5F ... merrily monkey occasionally odd puppy rabbit runs stupid swims weeps
0 -0.585248 0 0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 0 0 0
1 0.569125 0 1 0 0 0 0 0 0 0 ... 1 0 0 1 0 0 0 0 1 0
2 -0.076040 0 0 0 0 1 0 0 1 1 ... 0 1 0 0 0 0 0 0 0 0
3 -0.098878 0 0 0 0 0 0 0 0 0 ... 0 0 0 1 0 0 1 0 0 0
4 0.952773 0 0 0 0 0 0 0 0 0 ... 0 0 0 1 0 0 0 0 0 0
[5 rows x 85 columns]
Just clone this repository, and execute:
python -m tests.sample
This will execute a test sample, for you to get what's going on:
Starting from this dataframe (len=100):
A B C D
0 0.745236 BBM7UP [TUT7RS, MOW92W, 9O6IX6, T70X4Z] donkey barfs dirty foolishly
1 0.484822 GPC8CL [BG05XJ, IORYVC, BX9UK5, ERT4PJ] girl hits clueless dutifully
2 0.673377 BK3OE7 [GPC8CL, GPC8CL, GPC8CL, BG05XJ] car eats clueless occasionally
3 0.462564 AEAIH6 [] car eats dirty crazily
4 -0.115847 T70X4Z [] girl hits adorable crazily
With these metrics:
{u'A': {u'column_type': u'numeric',
u'mean': 0.048246178299744653,
u'std': 0.95162789611877563},
u'B': {u'column_type': u'mono-label',
u'imbalance_ratio': 6,
u'labels': 30,
u'occurrence_max': 6,
u'occurrence_mean': 3.3333333333333335,
u'occurrence_min': 1,
u'occurrence_std_dev': 1.4452988925785868},
u'C': {u'cardinality_mean': 1.6499999999999999,
u'cardinality_std_dev': 1.3955285736952863,
u'column_type': u'multi-label',
u'imbalance_ratio': 5,
u'labels': 29,
u'occurrence_max': 10,
u'occurrence_mean': 5.6896551724137927,
u'occurrence_min': 2,
u'occurrence_std_dev': 2.1189367580327199,
u'partitions': {u'imbalance_ratio': 29,
u'labels': 65,
u'occurrence_max': 29,
u'occurrence_mean': 1.5384615384615385,
u'occurrence_min': 1,
u'occurrence_std_dev': 3.4555503761871074}},
u'D': {u'column_type': u'mono-label',
u'imbalance_ratio': 2,
u'labels': 98,
u'occurrence_max': 2,
u'occurrence_mean': 1.0204081632653061,
u'occurrence_min': 1,
u'occurrence_std_dev': 0.14139190265868387}}
Filtering the small occurrences label of columns B and C...
We now get this (len=100):
A B C D
0 0.745236 BBM7UP [MOW92W, 9O6IX6] donkey barfs dirty foolishly
1 0.484822 GPC8CL [BG05XJ, IORYVC, ERT4PJ] girl hits clueless dutifully
2 0.673377 BK3OE7 [GPC8CL, GPC8CL, GPC8CL, BG05XJ] car eats clueless occasionally
3 0.462564 AEAIH6 [] car eats dirty crazily
4 -0.115847 T70X4Z [] girl hits adorable crazily
Filtering empty entry example for column B or C...
We finally have a clean dataframe (len=47):
A B C D
0 0.745236 BBM7UP [MOW92W, 9O6IX6] donkey barfs dirty foolishly
1 0.484822 GPC8CL [BG05XJ, IORYVC, ERT4PJ] girl hits clueless dutifully
2 0.673377 BK3OE7 [GPC8CL, GPC8CL, GPC8CL, BG05XJ] car eats clueless occasionally
5 -0.941320 7A37D6 [76AYX1] girl eats clueless dutifully
6 0.043402 7A37D6 [BK3OE7] rabbit jumps stupid merrily
With these metrics:
{u'A': {u'column_type': u'numeric',
u'mean': 0.14122463494338683,
u'std': 0.9745937014876852},
u'B': {u'column_type': u'mono-label',
u'imbalance_ratio': 5,
u'labels': 20,
u'occurrence_max': 5,
u'occurrence_mean': 2.3500000000000001,
u'occurrence_min': 1,
u'occurrence_std_dev': 1.0618380290797651},
u'C': {u'cardinality_mean': 1.7872340425531914,
u'cardinality_std_dev': 0.84893350396851086,
u'column_type': u'multi-label',
u'imbalance_ratio': 3,
u'labels': 15,
u'occurrence_max': 9,
u'occurrence_mean': 5.5999999999999996,
u'occurrence_min': 3,
u'occurrence_std_dev': 1.5405626677721789,
u'partitions': {u'imbalance_ratio': 4,
u'labels': 36,
u'occurrence_max': 4,
u'occurrence_mean': 1.3055555555555556,
u'occurrence_min': 1,
u'occurrence_std_dev': 0.69997795379745287}},
u'D': {u'column_type': u'mono-label',
u'imbalance_ratio': 1,
u'labels': 47,
u'occurrence_max': 1,
u'occurrence_mean': 1.0,
u'occurrence_min': 1,
u'occurrence_std_dev': 0.0}}