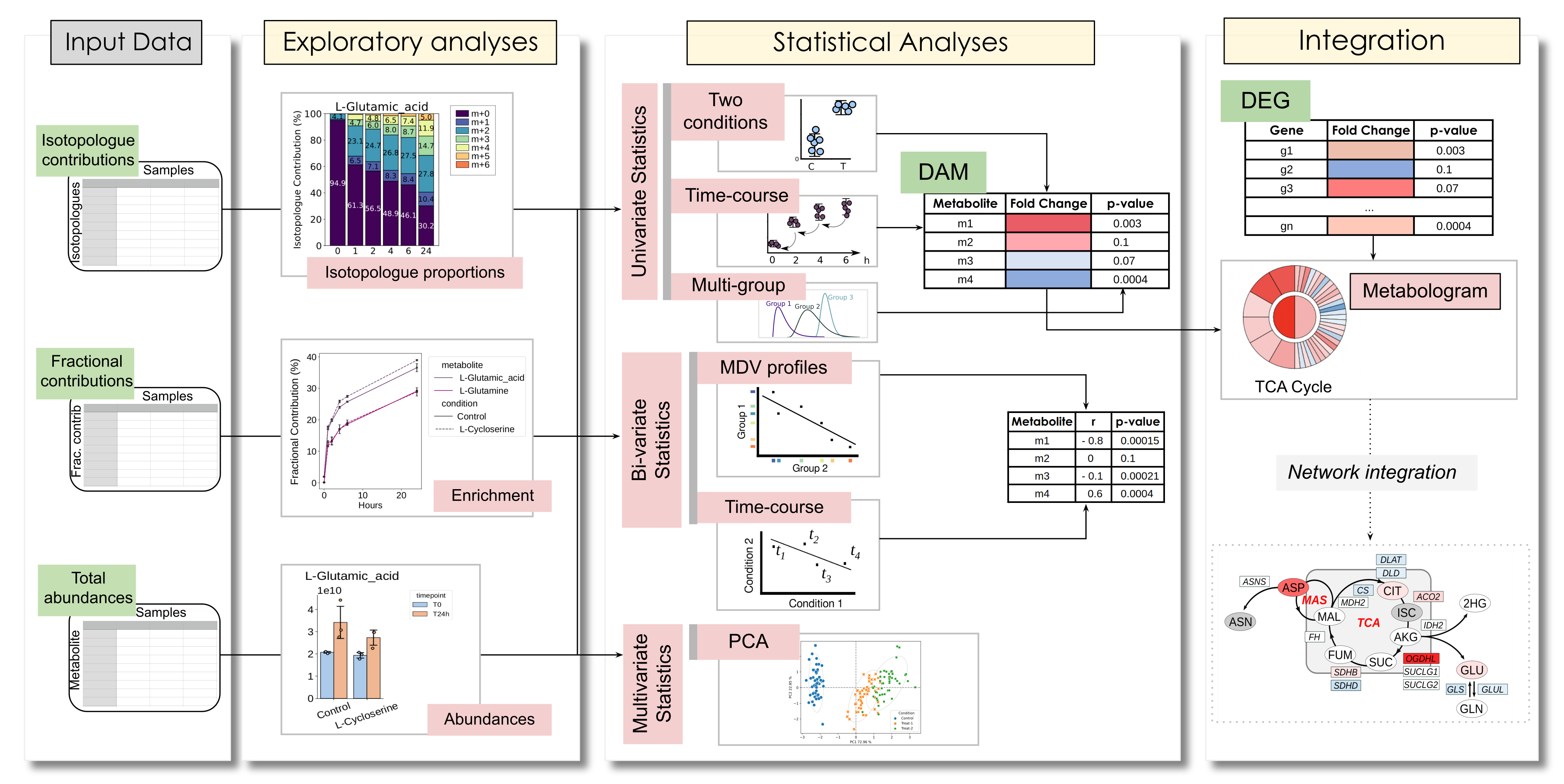
DIMet is a bioinformatics pipeline for differential and time-course analysis of targeted isotope-labeled metabolomics data.
DIMet supports the analysis of full metabolite abundances and isotopologue contributions, and allows to perform it in the differential comparison mode, or as a time-series analysis, or even processing entire labelling profiles. As input, DIMet accepts three types of measures: a) isotopologues’ contributions, b) fractional contributions (also known as mean enrichment), c) full metabolites’ abundances. DIMet also offers a pathway-based omics integration through Metabolograms.
DIMet functionalities (Galvis J., et al. Bioinformatics, 2024).Note: DIMet is intended for downstream analysis of tracer metabolomics data that has been corrected for the presence of natural isotopologues.
Formatting and normalisation helper: scripts for formatting and normalization are provided in TraceGroomer.
DIMet is available in Galaxy Europe (https://usegalaxy.eu/) and Galaxy Workflow4Metabolomics (https://workflow4metabolomics.usegalaxy.fr/), providing free access to the user-friendly interface of our tool.
Important
When using DIMet, please cite:
Galvis J, Guyon J, Dartigues B, Hecht H, Grüning B, Specque F, Soueidan H, Karkar S, Daubon T, Nikolski M. DIMet: An open-source tool for Differential analysis of targeted Isotope-labeled Metabolomics data. Bioinformatics 2024; btae282. https://doi.org/10.1093/bioinformatics/btae282
DIMet installation requires an Unix environment with python 3.9. It was tested under Linux and MacOS environments.
The full installation process should take less than 15 minutes on a standard computer.
Via pip command:
pip install dimet
Or if you are a developer working in a local cloned version, you can install:
pip install -e .
Alternatively to the PyPI version, our tool is also available as a conda package. Moreover, it can be used via Docker (docker pull quay.io/biocontainers/dimet:0.1.4) or singularity (depot.galaxyproject.org/singularity/dimet:0.1.4--pyhdfd78af_0) containers.
To start contributing to DIMet it is required to have a python environment with python >= 3.9 and poetry installed.
After creating the environment, the project can be installed with
poetry installsrc/dimet/processingdirectory contains the implemented high-level analysis scripts that produced the tables for the DIMet papersrc/dimet/visualization: directory contains the implemented high-level scripts that produced the figures in the DIMet papersrc/dimet/datadirectory contains the python classes for data initializationsrc/dimet/methoddirectory contains the python classes for configuration handlingtestsdirectory contains unit teststoolsdirectory contains venv setup scripts.
- With pytest, by running
pytestfromDIMet - Alternatively, place yourself in
DIMet/testsand executepython -m unittest - If the project was installed with
poetry install, tests can also be run usingpoetry run pytestor from VSCode's GUI
All the details about how to run DIMet can be found on the dedicated Wiki page. Importantly, this is where you will find the information about how to organise the data (folder structure) and how to populate the configuration files to successfully run DIMet.
For any information or help running DIMet, you can get in touch with:
Copyright (c) 2023
Johanna Galvis (1,2) deisy-johanna.galvis-rodriguez@u-bordeaux.fr
Benjamin Dartigues (2) benjamin.dartigues@u-bordeaux.fr
Florian Specque (1,2) florian.specque@u-bordeaux.fr
Slim Karkar (1,2) slim.karkar@u-bordeaux.fr
Helge Hecht (3,5) helge.hecht@recetox.muni.cz
Bjorn Gruening (4,5) bjoern.gruening@gmail.com
Hayssam Soueidan (2) massyah@gmail.com
Macha Nikolski (1,2) macha.nikolski@u-bordeaux.fr
(1) CNRS, IBGC - University of Bordeaux,
1, rue Camille Saint-Saens, Bordeaux, France
(2) CBiB - University of Bordeaux,
146, rue Leo Saignat, Bordeaux, France
(3) RECETOX
Faculty of Science, Masaryk University, Kotlářksá 2, 611 37 Brno, Czech Republic
(4) University of Freiburg,
Freiburg, Germany
(5) Galaxy Europe
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.