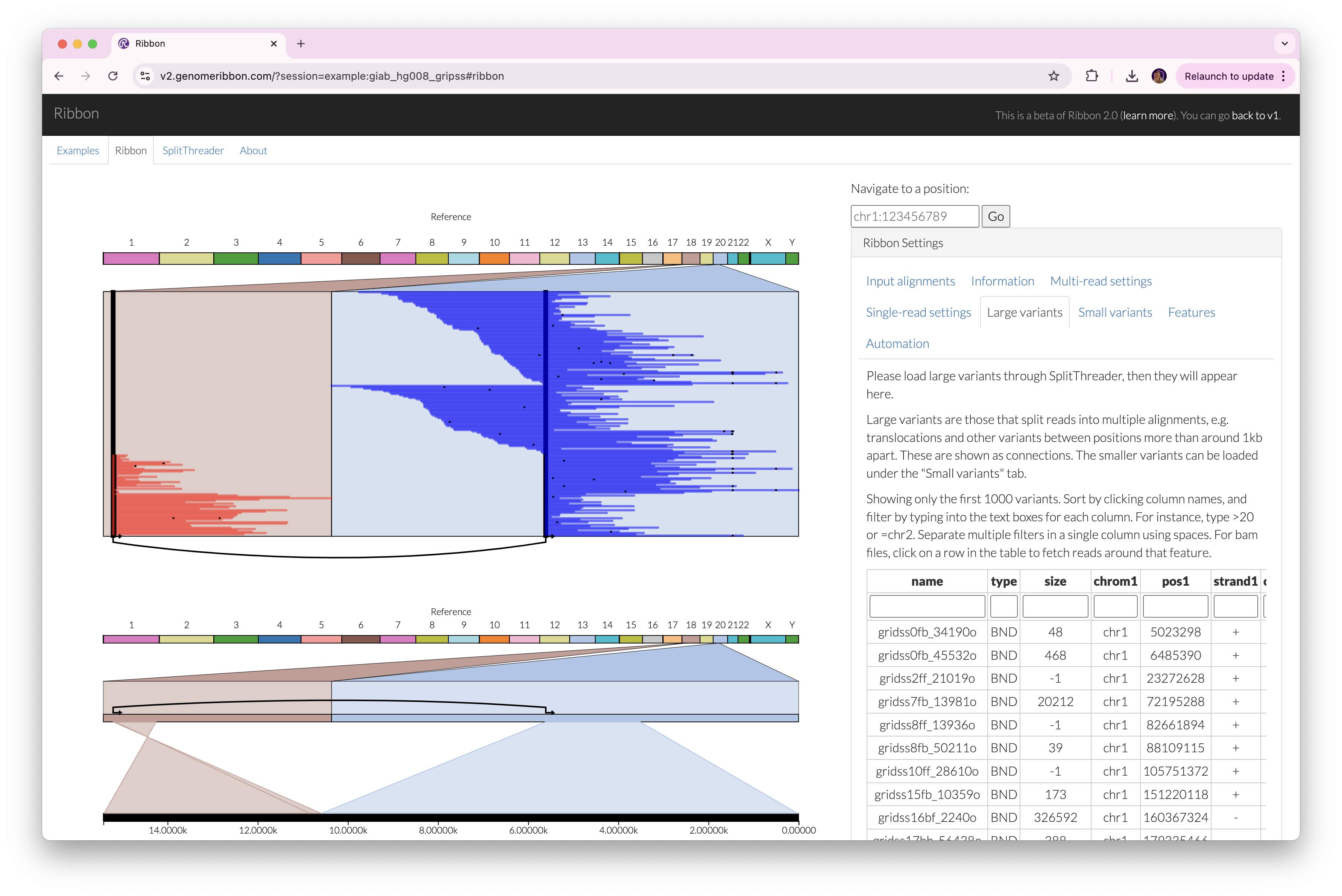
Ribbon and SplitThreader are both visualization tools for exploring complex rearrangements and long-range structural variants. Especially if you have long reads, Ribbon and SplitThreader should be quite helpful for figuring out what's going on around large variants.
Ribbon is available online at genomeribbon.com
Both built by Maria Nattestad originally, we decided in 2024 to put Ribbon and SplitThreader together and give them both major updates. This was prompted by a collaboration with the Genome in a Bottle (GIAB) team at NIST to support the release of their somatic (tumor) benchmark dataset for HG008. 2024 development done by Maria Nattestad and Robert Aboukhalil.
- How to read the visualizations? Check out interpretation for examples.
- What changed in 2024? Including how to make a session with multiple files: v2.0 release notes
Please cite our paper in Bioinformatics.
The BioRxiv has both the Ribbon preprint and the SplitThreader preprint.
There are several algorithms available within SplitThreader that can help make sense of long-range rearrangements, from categorizing variants:
 to searching for gene fusions:
gene fusion algorithms described in supplement to SplitThreader preprint.
to searching for gene fusions:
gene fusion algorithms described in supplement to SplitThreader preprint.
If you need to run Ribbon/SplitThreader without internet access, go to releases and find the ribbon_splitthreader.zip bundle from the latest release.
unzip ribbon_splitthreader.zip
cd ribbon_splitthreader
python3 -m http.server
# or run any other server pointed at that directory.Note that permalinks won't work without internet access.
To develop new features in Ribbon, first install npm.
Then clone this repository and build it:
# Clone repo and install dependencies
git clone https://github.com/marianattestad/ribbon
cd ribbon
npm install
# to run locally for development
npm run dev
# to build from source:
npm run build
npm run serve # to test the build
# To run a server with this build:
cd dist
python3 -m http.server