SDHB
| SDHB | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | SDHB, CWS2, IP, PGL4, SDH, SDH1, SDH2, SDHIP, succinate dehydrogenase complex iron sulfur subunit B, MC2DN4 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 185470; MGI: 1914930; HomoloGene: 2255; GeneCards: SDHB; OMA:SDHB - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial (SDHB) also known as iron-sulfur subunit of complex II (Ip) is a protein that in humans is encoded by the SDHB gene.[5][6][7]
The succinate dehydrogenase (also called SDH or Complex II) protein complex catalyzes the oxidation of succinate (succinate + ubiquinone => fumarate + ubiquinol). SDHB is one of four protein subunits forming succinate dehydrogenase, the other three being SDHA, SDHC and SDHD. The SDHB subunit is connected to the SDHA subunit on the hydrophilic, catalytic end of the SDH complex. It is also connected to the SDHC/SDHD subunits on the hydrophobic end of the complex anchored in the mitochondrial membrane. The subunit is an iron-sulfur protein with three iron-sulfur clusters. It weighs 30 kDa.
Structure
[edit]The gene that codes for the SDHB protein is nuclear, not mitochondrial DNA. However, the expressed protein is located in the inner membrane of the mitochondria. The location of the gene in humans is on the first chromosome at locus p36.1-p35. The gene is coded in 1,162 base pairs, partitioned in 8 exons.[5] The expressed protein weighs 31.6 kDa and is composed of 280 amino acids.[8][9] SDHB contains the iron-sulphur clusters necessary for tunneling electrons through the complex. It is located between SDHA and the two transmembrane subunits SDHC and SDHD.[10]
Function
[edit]
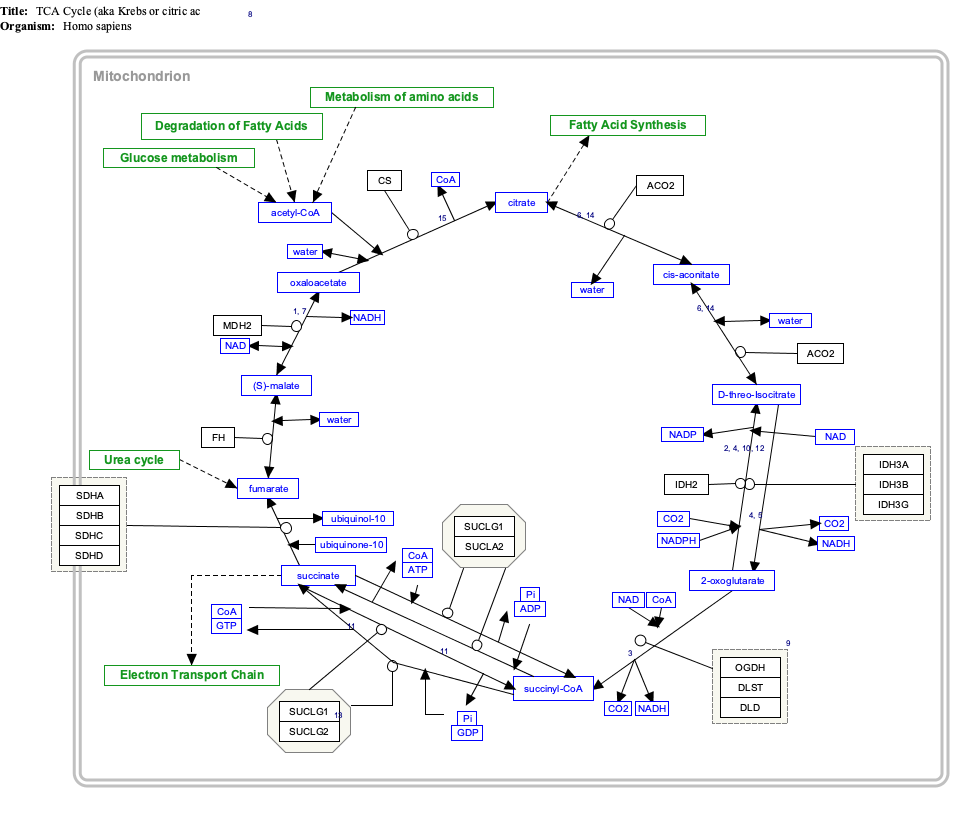
The SDH complex is located on the inner membrane of the mitochondria and participates in both the Citric Acid Cycle and Respiratory chain. SDHB acts as an intermediate in the basic SDH enzyme action shown in Figure 1:
- SDHA converts succinate to fumarate as part of the Citric Acid Cycle. This reaction also converts FAD to FADH2.
- Electrons from the FADH2 are transferred to the SDHB subunit iron clusters [2Fe-2S],[4Fe-4S],[3Fe-4S].
- Finally the electrons are transferred to the Ubiquinone (Q) pool via the SDHC/SDHD subunits. This function is part of the Respiratory chain.
Initially, SDHA oxidizes succinate via deprotonation at the FAD binding site, forming FADH2 and leaving fumarate, loosely bound to the active site, free to exit the protein. Electrons from FADH2 are transferred to the SDHB subunit iron clusters [2Fe-2S],[4Fe-4S],[3Fe-4S] and tunnel along the [Fe-S] relay until they reach the [3Fe-4S] iron sulfur cluster. The electrons are then transferred to an awaiting ubiquinone molecule at the Q pool active site in the SDHC/SDHD dimer. The O1 carbonyl oxygen of ubiquinone is oriented at the active site (image 4) by hydrogen bond interactions with Tyr83 of SDHD. The presence of electrons in the [3Fe-4S] iron sulphur cluster induces the movement of ubiquinone into a second orientation. This facilitates a second hydrogen bond interaction between the O4 carbonyl group of ubiquinone and Ser27 of SDHC. Following the first single electron reduction step, a semiquinone radical species is formed. The second electron arrives from the [3Fe-4S] cluster to provide full reduction of the ubiquinone to ubiquinol.[11]
Clinical significance
[edit]Germline mutations in the gene can cause familial paraganglioma (in old nomenclature, Paraganglioma Type PGL4). The same condition is often called familial pheochromocytoma. Less frequently, renal cell carcinoma can be caused by this mutation.
Paragangliomas related to SDHB mutations have a high rate of malignancy. When malignant, treatment is currently the same as for any malignant paraganglioma/pheochromocytoma.
Cancer
[edit]Paragangliomas caused by SDHB mutations have several distinguishing characteristics:
- Malignancy is common, ranging from 38%-83%[12][13] in carriers with disease. In contrast, tumors caused by SDHD mutations are almost always benign. Sporadic paragangliomas are malignant in less than 10% of cases.
- Malignant paragangliomas caused by SDHB are usually (perhaps 92%[13]) extra-adrenal. Sporadic pheochromocytomas/paragangliomas are extra-adrenal in less than 10% of cases.
- The penetrance of the gene is often reported as 77% by age 50[12] (i.e. 77% of carriers will have at least one tumour by the age of 50). This is likely an overestimate. Currently (2011), families with silent SDHB mutations are being screened[14] to determine the frequency of silent carriers.
- The average age of onset is approximately the same for SDHB vs non-SDHB related disease (approximately 36 years).
Mutations causing disease have been seen in exons 1 through 7, but not 8. As with the SDHC and SDHD genes, SDHB is a tumor suppressor gene.
Tumor formation generally follows the Knudson "two hit" hypothesis. The first copy of the gene is mutated in all cells, however the second copy functions normally. When the second copy mutates in a certain cell due to a random event, Loss of Heterozygosity (LOH) occurs and the SDHB protein is no longer produced. Tumor formation then becomes possible.
Given the fundamental nature of the SDH protein in all cellular function, it is not currently understood why only paraganglionic cells are affected. However, the sensitivity of these cells to oxygen levels may play a role.
Disease pathways
[edit]The precise pathway leading from SDHB mutation to tumorigenesis is not determined; there are several proposed mechanisms.[15]
Generation of reactive oxygen species
[edit]
When succinate-ubiquinone activity is inhibited, electrons that would normally transfer through the SDHB subunit to the Ubiquinone pool are instead transferred to O2 to create Reactive Oxygen Species (ROS) such as superoxide. The dashed red arrow in Figure 2 shows this. ROS accumulate and stabilize the production of HIF1-α. HIF1-α combines with HIF1-β to form the stable HIF heterodimeric complex, in turn leading to the induction of antiapoptotic genes in the cell nucleus.
Succinate accumulation in the cytosol
[edit]SDH inactivation can block the oxidation of succinate, starting a cascade of reactions:
- The succinate accumulated in the mitochondrial matrix diffuses through the inner and outer mitochondrial membranes to the cytosol (purple dashed arrows in Figure 2).
- Under normal cellular function, HIF1-α in the cytosol is quickly hydroxylated by prolyl hydroxylase (PHD), shown with the light blue arrow. This process is blocked by the accumulated succinate.
- HIF1-α stabilizes and passes to the cell nucleus (orange arrow) where it combines with HIF1-β to form an active HIF complex that induces the expression of tumor causing genes.[16]
This pathway raises the possibility of a therapeutic treatment. The build-up of succinate inhibits PHD activity. PHD action normally requires oxygen and alpha-ketoglutarate as cosubstrates and ferrous iron and ascorbate as cofactors. Succinate competes with α-ketoglutarate in binding to the PHD enzyme. Therefore, increasing α-ketoglutarate levels can offset the effect of succinate accumulation.
Normal α-ketoglutarate does not permeate cell walls efficiently, and it is necessary to create a cell permeating derivative (e.g. α-ketoglutarate esters). In-vitro trials show this supplementation approach can reduce HIF1-α levels, and may result in a therapeutic approach to tumours resulting from SDH deficiency.[17]
Impaired developmental apoptosis
[edit]Paraganglionic tissue is derived from the neural crest cells present in an embryo. Abdominal extra-adrenal paraganglionic cells secrete catecholamines that play an important role in fetal development. After birth these cells usually die, a process that is triggered by a decline in nerve growth factor (NGF)which initiates apoptosis (cell death).
This cell death process is mediated by an enzyme called prolyl hydroxylase EglN3. Succinate accumulation caused by SDH inactivation inhibits the prolyl hydroxylase EglN3.[18] The net result is that paranglionic tissue that would normally die after birth remains, and this tissue may be able to trigger paraganglioma/pheochromocytoma later.
Glycolysis upregulation
[edit]Inhibition of the Citric Acid Cycle forces the cell to create ATP glycolytically in order to generate its required energy. The induced glycolytic enzymes could potentially block cell apoptosis.
RNA editing
[edit]The mRNA transcripts of the SDHB gene in human are edited through an unknown mechanism at ORF nucleotide position 136 causing the conversion of C to U and thus generating a stop codon resulting in the translation of the edited transcripts to a truncated SDHB protein with an R46X amino acid change. This editing has been shown in monocytes and some human lymphoid cell-lines,[19] and is enhanced by hypoxia.[20]
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78".
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000117118 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000009863 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "Entrez Gene: succinate dehydrogenase complex".
- ^ Kita K, Oya H, Gennis RB, Ackrell BA, Kasahara M (January 1990). "Human complex II (succinate-ubiquinone oxidoreductase): cDNA cloning of iron sulfur (Ip) subunit of liver mitochondria". Biochem. Biophys. Res. Commun. 166 (1): 101–8. doi:10.1016/0006-291X(90)91916-G. PMID 2302193.
- ^ Au HC, Ream-Robinson D, Bellew LA, Broomfield PL, Saghbini M, Scheffler IE (July 1995). "Structural organization of the gene encoding the human iron-sulfur subunit of succinate dehydrogenase". Gene. 159 (2): 249–53. doi:10.1016/0378-1119(95)00162-Y. PMID 7622059.
- ^ Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, Deng N, Kim AK, Choi JH, Zelaya I, Liem D, Meyer D, Odeberg J, Fang C, Lu HJ, Xu T, Weiss J, Duan H, Uhlen M, Yates JR, Apweiler R, Ge J, Hermjakob H, Ping P (Oct 2013). "Integration of cardiac proteome biology and medicine by a specialized knowledgebase". Circulation Research. 113 (9): 1043–53. doi:10.1161/CIRCRESAHA.113.301151. PMC 4076475. PMID 23965338.
- ^ "SDHB - Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial". Cardiac Organellar Protein Atlas Knowledgebase (COPaKB). Archived from the original on 2018-07-19. Retrieved 2018-07-18.
- ^ Sun, F; Huo, X; Zhai, Y; Wang, A; Xu, J; Su, D; Bartlam, M; Rao, Z (1 July 2005). "Crystal structure of mitochondrial respiratory membrane protein complex II". Cell. 121 (7): 1043–57. doi:10.1016/j.cell.2005.05.025. PMID 15989954. S2CID 16697879.
- ^ Horsefield, R; Yankovskaya, V; Sexton, G; Whittingham, W; Shiomi, K; Omura, S; Byrne, B; Cecchini, G; Iwata, S (17 March 2006). "Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction". The Journal of Biological Chemistry. 281 (11): 7309–16. doi:10.1074/jbc.m508173200. PMID 16407191.
- ^ a b Neumann HP, Pawlu C, Peczkowska M, Bausch B, McWhinney SR, Muresan M, Buchta M, Franke G, Klisch J, Bley TA, Hoegerle S, Boedeker CC, Opocher G, Schipper J, Januszewicz A, Eng C (August 2004). "Distinct clinical features of paraganglioma syndromes associated with SDHB and SDHD gene mutations". JAMA. 292 (8): 943–51. doi:10.1001/jama.292.8.943. PMID 15328326. S2CID 21673619.
- ^ a b Brouwers FM, Eisenhofer G, Tao JJ, Kant JA, Adams KT, Linehan WM, Pacak K (November 2006). "High frequency of SDHB germline mutations in patients with malignant catecholamine-producing paragangliomas: implications for genetic testing". J. Clin. Endocrinol. Metab. 91 (11): 4505–9. doi:10.1210/jc.2006-0423. PMID 16912137.
- ^ Conference: National Institute of Health (U.S.A.), "SDHB-related Pheochromocytoma: Recent Discoveries & Current Diagnostic and Therapeutic Approaches", September 29, 2006
- ^ Gottlieb E, Tomlinson IP (November 2005). "Mitochondrial tumour suppressors: a genetic and biochemical update". Nat. Rev. Cancer. 5 (11): 857–66. doi:10.1038/nrc1737. PMID 16327764. S2CID 20851047.
- ^ Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E (January 2005). "Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase". Cancer Cell. 7 (1): 77–85. doi:10.1016/j.ccr.2004.11.022. PMID 15652751.
- ^ MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, Watson DG, Gottlieb E (May 2007). "Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells". Mol. Cell. Biol. 27 (9): 3282–9. doi:10.1128/MCB.01927-06. PMC 1899954. PMID 17325041.
- ^ Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, Farese RV, Freeman RS, Carter BD, Kaelin WG, Schlisio S (August 2005). "Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer". Cancer Cell. 8 (2): 155–67. doi:10.1016/j.ccr.2005.06.015. PMID 16098468.
- ^ Baysal BE (2007). "A recurrent stop-codon mutation in succinate dehydrogenase subunit B gene in normal peripheral blood and childhood T-cell acute leukemia". PLOS ONE. 2 (5): e436. Bibcode:2007PLoSO...2..436B. doi:10.1371/journal.pone.0000436. PMC 1855983. PMID 17487275.

- ^ Baysal BE, De Jong K, Liu B, Wang J, Patnaik SK, Wallace PK, Taggart RT (2013). "Hypoxia-inducible C-to-U coding RNA editing downregulates SDHB in monocytes". PeerJ. 1: e152. doi:10.7717/peerj.152. PMC 3775634. PMID 24058882.
Further reading
[edit]- Milosevic D, Lundquist P, Cradic K, et al. (2010). "Development and validation of a comprehensive mutation and deletion detection assay for SDHB, SDHC, and SDHD". Clin. Biochem. 43 (7–8): 700–4. doi:10.1016/j.clinbiochem.2010.01.016. PMC 3419008. PMID 20153743.
- Alrashdi I, Bano G, Maher ER, Hodgson SV (2010). "Carney triad versus Carney Stratakis syndrome: two cases which illustrate the difficulty in distinguishing between these conditions in individual patients". Fam. Cancer. 9 (3): 443–7. doi:10.1007/s10689-010-9323-z. PMID 20119652. S2CID 21792188.
- Okada Y, Kamatani Y, Takahashi A, et al. (2010). "A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci". Hum. Mol. Genet. 19 (11): 2303–12. doi:10.1093/hmg/ddq091. PMID 20189936.
- Bayley JP (2010). "Are these compound heterozygous mutations of SDHB really mutations?". Pediatr Blood Cancer. 55 (1): 211, author reply 212. doi:10.1002/pbc.22455. PMID 20213850. S2CID 31378042.
- Rose JE, Behm FM, Drgon T, et al. (2010). "Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score". Mol. Med. 16 (7–8): 247–53. doi:10.2119/molmed.2009.00159. PMC 2896464. PMID 20379614.
- Gill AJ, Benn DE, Chou A, et al. (2010). "Immunohistochemistry for SDHB triages genetic testing of SDHB, SDHC, and SDHD in paraganglioma-pheochromocytoma syndromes". Hum. Pathol. 41 (6): 805–14. doi:10.1016/j.humpath.2009.12.005. PMID 20236688.
- Martin TP, Irving RM, Maher ER (2007). "The genetics of paragangliomas: a review". Clin Otolaryngol. 32 (1): 7–11. doi:10.1111/j.1365-2273.2007.01378.x. PMID 17298303.
- Eng C, Kiuru M, Fernandez MJ, Aaltonen LA (2003). "A role for mitochondrial enzymes in inherited neoplasia and beyond". Nat. Rev. Cancer. 3 (3): 193–202. doi:10.1038/nrc1013. PMID 12612654. S2CID 20549458.
- Lee J, Wang J, Torbenson M, et al. (2010). "Loss of SDHB and NF1 genes in a malignant phyllodes tumor of the breast as detected by oligo-array comparative genomic hybridization". Cancer Genet. Cytogenet. 196 (2): 179–83. doi:10.1016/j.cancergencyto.2009.09.005. PMID 20082856.
- Hermsen MA, Sevilla MA, Llorente JL, et al. (2010). "Relevance of germline mutation screening in both familial and sporadic head and neck paraganglioma for early diagnosis and clinical management". Cell. Oncol. 32 (4): 275–83. doi:10.3233/CLO-2009-0498. PMC 4619289. PMID 20208144.
- Musil Z; Puchmajerová A; Krepelová A; et al. (2010). "Paraganglioma in a 13-year-old girl: a novel SDHB gene mutation in the family?". Cancer Genet. Cytogenet. 197 (2): 189–92. doi:10.1016/j.cancergencyto.2009.11.010. PMID 20193854.
- Shimada M, Miyagawa T, Kawashima M, et al. (2010). "An approach based on a genome-wide association study reveals candidate loci for narcolepsy". Hum. Genet. 128 (4): 433–41. doi:10.1007/s00439-010-0862-z. PMID 20677014. S2CID 24207887.
- Brière JJ; Favier J; El Ghouzzi V; et al. (2005). "Succinate dehydrogenase deficiency in human". Cell. Mol. Life Sci. 62 (19–20): 2317–24. doi:10.1007/s00018-005-5237-6. PMC 11139140. PMID 16143825. S2CID 23793565.
- Schimke RN, Collins DL, Stolle CA (2010). "Paraganglioma, neuroblastoma, and a SDHB mutation: Resolution of a 30-year-old mystery". Am. J. Med. Genet. A. 152A (6): 1531–5. doi:10.1002/ajmg.a.33384. PMID 20503330. S2CID 22768946.
- Gill AJ, Chou A, Vilain R, et al. (2010). "Immunohistochemistry for SDHB divides gastrointestinal stromal tumors (GISTs) into 2 distinct types". Am. J. Surg. Pathol. 34 (5): 636–44. doi:10.1097/PAS.0b013e3181d6150d. PMID 20305538. S2CID 2314622.
- Hendrickson SL, Lautenberger JA, Chinn LW, et al. (2010). "Genetic variants in nuclear-encoded mitochondrial genes influence AIDS progression". PLOS ONE. 5 (9): e12862. Bibcode:2010PLoSO...512862H. doi:10.1371/journal.pone.0012862. PMC 2943476. PMID 20877624.

- Cerecer-Gil NY, Figuera LE, Llamas FJ, et al. (2010). "Mutation of SDHB is a cause of hypoxia-related high-altitude paraganglioma". Clin. Cancer Res. 16 (16): 4148–54. doi:10.1158/1078-0432.CCR-10-0637. PMID 20592014. S2CID 12502978.
- Krawczyk A, Hasse-Lazar K, Pawlaczek A, et al. (2010). "Germinal mutations of RET, SDHB, SDHD, and VHL genes in patients with apparently sporadic pheochromocytomas and paragangliomas". Endokrynol Pol. 61 (1): 43–8. PMID 20205103.
- Hes FJ, Weiss MM, Woortman SA, et al. (2010). "Low penetrance of a SDHB mutation in a large Dutch paraganglioma family". BMC Med. Genet. 11: 92. doi:10.1186/1471-2350-11-92. PMC 2891715. PMID 20540712.

- Bailey SD, Xie C, Do R, et al. (2010). "Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study". Diabetes Care. 33 (10): 2250–3. doi:10.2337/dc10-0452. PMC 2945168. PMID 20628086.
- Baysal BE (2007). "A Recurrent Stop-Codon Mutation in Succinate Dehydrogenase Subunit B Gene in Normal Peripheral Blood and Childhood T-Cell Acute Leukemia". PLOS ONE. 2 (5): e436. Bibcode:2007PLoSO...2..436B. doi:10.1371/journal.pone.0000436. PMC 1855983. PMID 17487275.

- Baysal BE, Jong KD, Liu B, et al. (2013). "Hypoxia-inducible C-to-U coding RNA editing downregulates SDHB in monocytes". PeerJ. 1: e152. doi:10.7717/peerj.152. PMC 3775634. PMID 24058882.
External links
[edit]- Database of identified SDHB mutations. Archived 2006-04-21 at the Wayback Machine