The Zeiformes (dories) are mid-water or deep (to 1000 m) marine acanthomorph fishes with a global, circumtropical, and circumtemperate distribution. Some species have a near-worldwide distribution, while others appear to be regional endemics, e.g., near New Zealand. Six families, 16 genera, and 33 species are currently recognized as valid. Relationships among them, however, remain unsettled, especially in light of recent proposals concerning the phylogenetic placement of zeiforms within the Paracanthopterygii rather than allied with beryciforms or percomorphs. The present study uses both morphological and molecular characters to investigate zeiform interrelationships given their revised phylogenetic placement and attendant changes to their close outgroups. Results indicate that revised outgroups affected the phylogenetic conclusions, especially those based on morphology. All analyses recovered monophyletic Zeidae, Cyttidae, and Oreosomatidae. Zeniontidae were recovered as polyphyletic, with the clade Capromimus Cyttomimus sister to Oreosomatidae. Based on morphological evidence, Grammicolepididae are paraphyletic. Parazenidae are monophyletic in all results except maximum likelihood based on molecular data. Morphometric analysis revealed a star-like radiation in morphospace with three diverging trends, each trend exemplified by convergences in body form. Overall, our results are suggestive of a rapid diversification among the major lineages of Zeiformes during the Late Cretaceous.
THE Zeiformes (i.e., dories, lookdown dories, tinselfishes, and oreos) are mid-water or deep (reported to 1,000 m) marine acanthomorph fishes with a global, circumtropical, and circumtemperate distribution. Some species apparently have a near-worldwide distribution, while others appear to be regional endemics (e.g., around New Zealand; Fig. 1). Zeiforms have a fossil record dating to the Late Cretaceous (late Campanian/early Maastrichtian, 72 mya; Tyler et al., 2000; Baciu et al., 2005; Tyler and Santini, 2005; Davesne et al., 2017). Thirty-three extant species are currently recognized as valid and are distributed among 16 genera in six families (Tyler et al., 2003; Tyler and Santini, 2005; Nelson et al., 2016). The relationships among the species, genera, and families may be in need of reexamination in light of recent proposals concerning the phylogenetic placement of zeiforms within the Paracanthopterygii rather than allied with beryciforms or percomorphs.
Fig. 1.
World map showing the collecting localities for specimens from which tissues were obtained for this study, along with the type locality for each species in the order. Specimen and type localities are most numerous in the western and southwestern Pacific, in the waters surrounding southern Africa, and on either side of the North Atlantic.

The question of the phylogenetic placement of zeiform fishes among acanthomorphs has a complicated and storied history. Various zeiform subgroups have been aligned with scombrids (Günther, 1860), chaetodontids and acanthuroids (Starks, 1898), pleuronectiforms (Holt, 1894; Boulenger, 1902), and caproiforms (Regan, 1910). Patterson (1968) aligned zeiforms + caproids with beryciforms. Rosen (1984) argued that the inclusion of caproids within zeiforms rendered Zeiformes non-monophyletic, and argued that zeiforms (excluding caproids) were a division within the order Tetraodontiformes. Johnson and Patterson (1993), in a morphological analysis of Acanthomorpha, noted some similarities in caudal-fin morphology (e.g., full spine on preural centrum 2, and a free ural centrum 2 during development) between zeiforms and percopsiforms. Although they removed zeiforms from Percomorpha, they did not group them with percopsiforms but placed them as sister to Beryciformes + Percomorpha.
Wiley et al. (2000) recovered, for the first time, a zeiform-gadiform sister group relationship in a total-evidence analysis of acanthomorphs incorporating the 34 morphological characters of Johnson and Patterson (1993) plus 1,674 base pairs from two ribosomal gene fragments (572 base pairs from mitochondrial 12S and 1,112 base pairs from nuclear 28S). When their data set was partitioned, however, only the molecular data supported the zeiform-gadiform relationship. Subsequent molecular analyses (e.g., Chen et al., 2003; Miya et al., 2003; Dettaï and Lecointre, 2005; Smith and Wheeler, 2006) continued to support a zeiform-gadiform relationship. The relationship of zeiforms to gadiforms has since been corroborated by broad-scale molecular phylogenetic analyses (e.g., Near et al., 2012; Betancur-R. et al., 2013; Chen et al., 2014). Another advance occurred when Miya et al. (2007), using mitochondrial data, recovered the supposed lampriform Stylephorus as sister to gadiforms, with zeiforms sister to Stylephorus + gadiforms. This zeiform-Stylephorus-gadiform relationship was also supported by Grande et al. (2013) using both molecular (nuclear and mitochondrial) and morphological data. In the latter study, zeiforms fell within Paracanthopterygii, which then consisted of percopsiforms + [zeiforms + (Stylephorus + gadiforms)]. The primitive acanthomorph genus Polymixia was recovered as sister to their Paracanthopterygii, and some authors now include Polymixia within an enlarged Paracanthopterygii (e.g., Nelson et al., 2016). Most recently, Davesne et al. (2016) used morphological data that supported a close relationship among zeiforms, Stylephorus, and gadiforms within their “Clade B,” which corresponds to the Paracanthopterygii (i.e., Polymixiiformes, Percopsiformes, Gadiformes, Zeiformes, Stylephorus) of Nelson et al. (2016). The phylogenetic position of zeiforms within Paracanthopterygii thus seems to be well established.
To date the most comprehensive analyses of zeiform intra-relationships are those of Tyler et al. (2003) and Tyler and Santini (2005). Those studies relied on morphology-based phylogenetic analyses for testing a tetraodontiform-zeiform sister-group relationship (the analysis of 2003 was rooted on Melamphaes suborbitalis, a beryciform); three of the four analyses in Tyler et al. (2003) supported that relationship, but not the one that they considered the most rational and best justified (unordered, without most meristics), which left those relationships unresolved (Tyler et al., 2003:fig. 5). Their analyses did not include a gadiform, a percopsiform, Stylephorus, or Polymixia, the four closest relatives of zeiforms according to current phylogenies (e.g., Grande et al., 2013; Davesne et al., 2016). In the results of Tyler et al. (2003), Cyttidae were sister to the remaining Zeiformes. This latter clade was divided into two, with Oreosomatidae in one and sister to a second clade consisting of Parazenidae + [Zeniontidae + (Grammicolepididae + Zeidae)].
Although fossil zeiforms were discussed by Tyler et al. (2003), fossil taxa were not included in a phylogenetic analysis with extant forms until the work of Tyler and Santini (2005). In the latter study, three fossils currently recognized as zeiforms were included in analyses: the Late Cretaceous (late Campanian/early Maastrichtian) †Cretazeus rinaldii from Italy (Tyler et al., 2000), as well as two small fossils from Denmark of late Paleocene/early Eocene (about 56 mya) age. The last two fossils have been called “†Protozeus kuehnei” and “†Archaeozeus skamolensis” and attributed to Bonde and Tyler in Tyler et al. (2003; e.g., by Tyler and Santini, 2005, and Baciu et al., 2005, receiving more complete descriptions in the latter publication), but unfortunately both of these generic names and their type species appear to us to be nomina nuda because they do not satisfy Article 16.1 of the most recent Code (ICZN, 1999), “intention of authors to establish new nominal taxa to be explicit.” All three fossils were added to the morphological matrix of Tyler et al. (2003), and the matrix was analyzed by Tyler and Santini (2005) under parsimony. In one of the Tyler and Santini (2005) analyses, using a reduced taxon set (all outgroups except two beryciforms, Melamphaes and Sargocentron, having been removed), Parazenidae were not recovered as monophyletic, with Parazen sister to Zeniontidae (sensu Tyler et al., 2003). †Cretazeus rinaldii was nested within more derived zeiforms as sister to Cyttopsis + Stethopristes. The two fossils from Denmark were recovered as sequential stem-group zeiforms.
The early Eocene fossil †Bajaichthys elegans Sorbini, 1983 was originally described as a zeiform, but Sorbini and Bottura (1988) later attributed it to the Lampriformes. Most recently, Davesne et al. (2017) have once again assigned it to the Zeiformes. Using morphological data they recovered a basal polytomy that included †Bajaichthys, but when they added the molecular data from Grande et al. (2013), they placed †Bajaichthys and the two fossils from Denmark as stem-group zeiforms.
The present study uses both morphological and molecular characters to focus on the genus- and family-level relationships within zeiforms in light of their revised phylogenetic position (i.e., within Paracanthopterygii), with the attendant changes to their close outgroups. Our use of multiple data sources and methods of analysis allows for comparative and independent assessments of phylogenetic relationships and of their implications.
The overall goal of this paper is to provide a better understanding of the evolution and relationships of the Zeiformes by asking the following questions: (1) Do the revised phylogenetic position of Zeiformes and the consequent changes in outgroup selection affect the proposed within-group relationships of Tyler et al. (2003) and Tyler and Santini (2005)? (2) Are phylogenetic inferences from DNA sequences congruent with those from morphology, and if not, what might account for those differences? (3) Are the constituent families as recognized by Tyler et al. (2003) and Tyler and Santini (2005) monophyletic based on the new analyses? (4) Are the major differences in body shapes of zeiforms the result of unique or convergent events?
MATERIALS AND METHODS
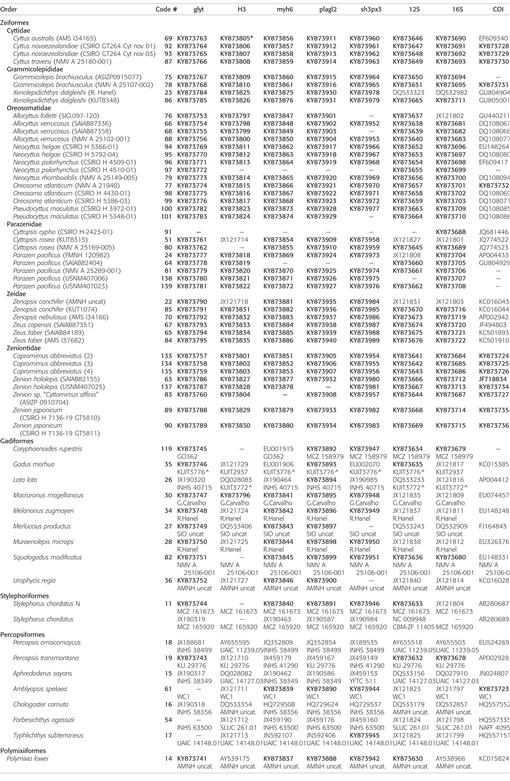
Molecular phylogenetic methods.—Our approach to taxonomic sampling relied on the ingroup phylogenetic hypotheses of Tyler et al. (2003) and Tyler and Santini (2005). We acquired at least 44 tissues (Table 1) representing all six nominal families (Cyttidae, Grammicolepididae, Oreosomatidae, Parazenidae, Zeidae, and Zeniontidae), including 13 of 16 genera (exceptions were Cyttomimus, Macrurocyttus, and Stethopristes, all of which were included in our morphological analysis), and 22 of the 33 nominal species (Table 1). Eight of the 11 missing species are represented by congenerics. The sequences used in this study, most of them new, represent eight loci (five nuclear and three mitochondrial). They are shown with GenBank accession numbers in Table 1. Within Zeiformes, we analyzed 374 sequences, with 290 (78%) of them new. For the outgroups, we analyzed 133 sequences, with 52 (39%) of them new. We built upon the sequence matrices of Grande et al. (2013) and Betancur-R. et al. (2013) to minimize the number of chimeric taxa. The sampled tissues came from all major oceans except the Arctic (Fig. 1).
Table 1.
List of tissues and sequences for each of eight loci used in this study, with voucher numbers where available. “12S” consists of a large portion of 12S rRNA gene, the complete tRNA-Val gene, and a fragment of the 16S rRNA gene. GenBank accession numbers in bold are new; other accession numbers are from earlier studies. Some new sequences differ from or are longer than sequences available from GenBank. * a shorter sequence by ≥30%; ^ tissue has no voucher.
Outgroups were selected based on recent molecular-based hypotheses (e.g., Near et al., 2012; Betancur-R. et al., 2013; Grande et al., 2013), all of which suggested that zeiforms are the sister group to gadiforms plus stylephoriforms and are more distantly related to percopsiforms and polymixiforms. The trees were rooted on the last common ancestor shared with Polymixia. Outgroups such as the beryciforms and tetraodontiforms, formerly considered to be closer relatives, were omitted from the new analyses.
DNA extraction, locus amplification, and sequence alignment.—Genomic DNA was extracted from ethyl alcohol-preserved material following guidelines of the DNeasy Blood and Tissue Kit (Qiagen). Three mitochondrial and five nuclear genes were amplified and sequenced. Primer sets and thermal cycler regimes for mitochondrial (12S: Titus, 1992; Feller and Hedges, 1998; 16S: Kocher et al., 1989; Palumbi, 1996; COI: Ward et al., 2005; Ivanova et al., 2007) and nuclear (H3: Colgan et al., 1998, glyt [gtdc2 in Betancur-R. et al., 2013], myh6, plagl2, sh3px3 [snx33 in Betancur-R. et al., 2013]: Li et al., 2007) loci were taken from the published literature. Loci glyt, myh6, plagl2, and sh3px3 were amplified using nested primer sets in which products from external amplifications were diluted and used as templates with internal primer sets (Li et al., 2007). Amplicons were sequenced by the University of Washington (Seattle, WA) High Throughput Genomics Center, and contigs were constructed and edited in Sequencher (Gene Codes) or Geneious v7.1.9 (Kearse et al., 2012; www.geneious.com). Failed amplifications from these methods were attempted using PCR beads in premixed and predispensed reaction tubes (puReTaq Ready-To-Go PCR Beads by illustra™).
Alignment of 12S and 16S sequences was performed with an online version of MAFFT v7.110 (Katoh and Toh, 2008) and the option “Q-INS-i”, which incorporates secondary structure of rDNA into the alignment algorithm. The resulting output was reviewed and edited by eye in Se-Al v.2.0a11 (Rambaut, 1996). Protein-coding loci were aligned either by eye after translation to amino acids when indels were absent or with an online version of Revtrans 1.4 (Wernersson and Pedersen, 2003) when indels were present (glyt, plagl2). The latter algorithm simultaneously considers nucleotide and amino acid inputs to construct an alignment.
To identify potential instances of cross-contamination and voucher misidentification, sequences were vetted by constructing gene trees using MrBayes v.3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) that also incorporated additional zeiform and outgroup sequences from online databases. Results of this vetting highlighted several questionable sequences and two misidentified vouchers (Cyttomimus ASIZP 0910740 and Neocyttus rhomboidalis SAIAB 87358), whose identifications were re-examined by staff at their home institutions. The revised identifications (Zenion sp. and Allocyttus verrucosus, respectively) matched those suggested by our gene trees. Suspect sequences were omitted, and new amplifications were re-sequenced, in some cases from new extractions. This process was repeated until suspicious sequences had been addressed. In several instances, this resulted in missing sequences.
A matrix of 64 terminals (43 zeiforms) totaling 5,387 bp (1,835 of them parsimony-informative) was constructed from eight loci (mtDNA—12S: 629 bp; 16S: 566 bp; COI: 654 bp; nDNA—glty: 870 bp; H3: 339 bp; myh6: 781 bp; plagl2: 810 bp; sh3px3: 738 bp), in which only 34 out of 512 sequences (∼6.5%) were missing, half of them being in loci H3 and sh3px3 (n = 8 and 9, respectively). Neocyttus psilorhynchus (sample 97) was missing five of eight sequences (Table 1) but was retained in the analyses; Cyttopsis cypho was missing six of eight and was removed from analyses. Matrices are available as supplemental material (see Data Accessibility).
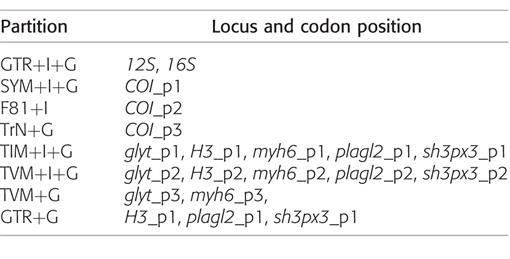
Partitioning schemes and nucleotide substitution models.—Biases of nucleotide frequencies among taxa, which can introduce systematic errors, were tested using the “basefreqs” command in PAUP* v4.0b10 (Swofford, 2003) by applying a chi-square test of homogeneity. Each locus was subjected to this test after removal of missing sequences and the p-value used as an indication of whether nucleotide frequencies varied significantly among taxa. All loci failed to show nucleotide frequency biases among taxa.
The most appropriate partition scheme with nucleotide substitution models was determined using a greedy algorithm and BIC criterion in PartitionFinder (Lanfear et al., 2012). The preferred scheme (Table 2) identified eight partitions for the molecular data, applied in all analyses.
Table 2.
Nucleotide substitution models applied to sequence partitions as determined by the greedy algorithm and BIC criterion in PartitionFinder (Lanfear et al., 2012) for the Maximum Likelihood and Bayesian Inference analyses.
Phylogenetic analysis of sequence matrices.—Maximum likelihood (ML) analyses were conducted using Garli v2.0 (Zwickl, 2006). To improve confidence that the analysis converged on the correct tree, two separate analyses of 100 search iterations were performed with taxa added in stepwise addition. Nodal support was estimated using a nonparametric bootstrap from 1,000 pseudoreplicates.
Bayesian analyses were performed with MrBayes v3.2.6 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). A Markov Chain Monte Carlo (MCMC) sampler ran for 25 × 106 generations using four chains and sampling frequency of 1/2000 generations. The mean exponential prior on branch lengths was decreased to 0.01 (default = 0.1) in order to minimize the possibility that runs would remain trapped in local minima (Brown et al., 2010; Marshall, 2010). Following a 25% burn-in, before which the –ln(likelihood) had stabilized, model parameters and sampled topologies were summarized, and a 50% majority rule consensus tree was constructed from sampled trees. Nodal confidence was indicated by posterior probabilities (PP). Finally, convergence of the two runs was assessed using the average standard deviation of split frequencies, plot of –lnL versus the number of generations, potential scale reduction factors (Gelman and Rubin, 1992), and the “compare” command in Are We There Yet (Wilgenbusch et al., 2004). Trees were visualized with FigTree v.1.4.2 (Rambaut, 2009).
In addition to analyses using our revised outgroups as described above, we also tested the effects of using the same outgroups as Tyler et al. (2003), none of which is now regarded as a close relative of Zeiformes.
Morphological phylogenetic methods.—A character matrix of 119 characters (Table 3) was assembled in Mesquite 3.2 (Maddison and Maddison, 2016). Characters 1–103 from Tyler and Santini (2005) made up the foundation of our data matrix (Appendix 1). Fifteen of their characters overlap with those of Grande et al. (2013) and are designated in Appendix 1. Characters 92 and 104–107 of Tyler and Santini (2005) were eliminated because they were invariant within our set of taxa and replaced with new characters from Grande et al. (2013) and Borden et al. (2013). To better understand how to code the proposed character states of Tyler and Santini (2005) in the (new) outgroups, it was necessary also to directly examine specimens of the zeiform taxa (see Material Examined). Characters 21, 40, 46, 53, 58, 60, 61, 70–72, 80, 81, 86–88, 94, 100, 101, and 103 from Tyler and Santini (2005) were rewritten or modified.
Table 3.
Morphological character matrix based on the data of Tyler and Santini (2005), with deletions, additions, and edits as described in the text. Multi-state characters are treated as polymorphisms.
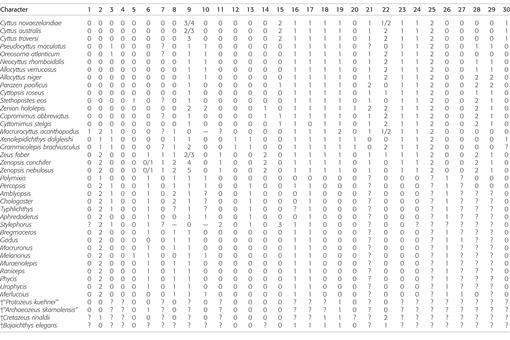
Table 3.
Continued.

Table 3.
Continued.
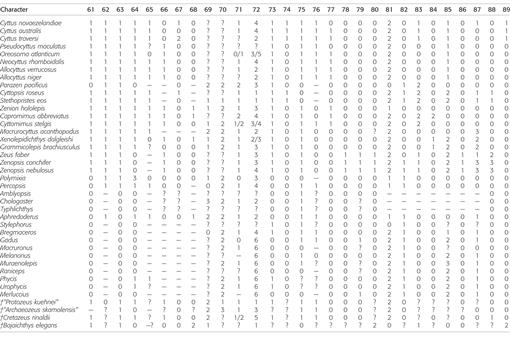
Table 3.
Continued.
The total number of ingroup taxa included in the morphological analysis was 20; these included all three of the genera (Cyttomimus, Macrurocyttus, and Stethopristes) and one species (Allocyttus niger) for which no molecular data were available. No specimen of the rare Macrurocyttus acanthopodus available to Tyler et al. (2003) was larger than 45 mm SL, within the usual size range for larval specimens of extant zeiforms, and many specimens were represented by disarticulated bones (Tyler et al., 2003:15); nevertheless, there were sufficient intact specimens along with the disarticulated bones to allow for relatively complete coding. Seven outgroup taxa (Polymixia, Percopsis, Chologaster, Muraenolepis, Urophycis, Merluccius, and Gadus) were ultimately chosen for the morphological analysis based on the desire to parallel the outgroups chosen for the molecular component of this study and the need to include outgroup taxa with the most complete character information. Although Stylephorus is represented in our character matrix, it was not included as an outgroup in our analysis because of the large number (42) of missing or inapplicable character states. For similar reasons, we did not include in our analyses some of the gadiform and percopsiform outgroups with large proportions of missing data, nor did we include in analyses the similarly incompletely coded fossil zeiforms (the coding of these by Tyler and Santini, 2005, is shown in Table 3). We estimate below the most parsimonious position of these fossils in a constrained phylogenetic topology.
The significant amount of missing data for these fossil taxa may be partly related to their small size and larval life-history features, making them not strictly comparable to adult specimens of extant taxa coded for in this study. All of these early fossil zeiforms are represented by very small individuals. The two fossil species from Denmark include specimens of 8.5 mm and 10.5 mm SL for “†Protozeus kuehnei” and 9.5 mm SL for the single known specimen of “†Archaeozeus skamolensis” (Tyler et al., 2005); for †Cretazeus rinaldii, known specimens range from 15–53 mm SL (Tyler et al., 2003), while for †Bajaichthys elegans, the single known specimen is 38.5 mm in SL (Davesne et al., 2017). Such small sizes correspond to post-flexion and metamorphic larval developmental stages of extant zeiforms (e.g., Kendall et al., 1984; Tighe and Keene, 1984; Ditty, 2003); therefore, their morphological features must be interpreted with caution, as recognized and discussed by Tyler et al. (2003:p. 5).
From our initial matrix of 119 characters (Table 3), 14 characters (4, 21, 24, 25, 27–29, 38, 47–48, 69, 70, 91, 95) were excluded before analysis, leaving 105 characters that were included. Characters were omitted in this study if they were phylogenetically uninformative, contained a significant number of “?” in the ingroup, or were judged to be overly subjective. Data were analyzed by the criterion of maximum parsimony with all characters unordered and unweighted, as in the methods used by Tyler and Santini (2005). Searching for the shortest tree was by the heuristic option in PAUP v.4b10 (Swofford, 2003) using the following settings: starting trees by stepwise addition with 1,000 random-addition sequence replicates and one tree held at each step, branch swapping by tree-bisection-reconnection (TBR) with reconnection limit eight, steepest descent not in effect, unlimited trees retained for swapping, mulTrees option in effect, and no topological constraints. Resulting trees were rooted on the outgroup Polymixia. For evaluation of the robustness of the results, both bootstrap (Felsenstein, 1985) and decay (Bremer, 1988) were calculated. One thousand bootstrap replicates were performed; for each bootstrap replicate, searching was heuristic with TBR and 100 random addition sequence replicates. Decay values were obtained by searching in PAUP with the same methods as the initial search except for finding the strict consensus of all trees shorter than [(shortest tree) + n], where the decay value n varied from one to nine. Clades with decay values greater than nine (indicated on the tree as ≥10) were not calculated because of excessive processing time and memory constraints. Character changes for the shortest tree were mapped onto the tree using MacClade 4.08 (Maddison and Maddison, 2005).
Analysis of a combined DNA sequence and morphological matrix.—Morphology and DNA sequences were combined in a matrix of 41 taxa and 5,492 characters (105 of them morphological characters). Multiple specimens of a given species were removed, and any taxon that had either DNA sequences or morphology was retained. In addition, the number of outgroups was reduced to 13 taxa (one Polymixia, five percopsiforms, Stylephorus, six gadiforms). The morphological matrix was considered its own partition and analyzed under the Mk model in Garli or the standard model (Lewis, 2001) in MrBayes. In Garli, 200 searches and 1,000 bootstrap replicates were performed. Bayesian analysis followed the parameters described earlier.
Morphometric methods.—Morphometric analyses were conducted to understand the variation in zeiform body shapes and the implications of the combined-evidence phylogeny for body-shape evolution. Twenty-eight images, one image per species, encompassing all valid extant zeiform genera and almost all species were assembled from new photographs taken of museum specimens and supplemented by images from well-curated and well-documented museum image databases (Material Examined). With one exception (Macrurocyttus acanthopodus), all images used were of large, juvenile or adult specimens with closed mouths and with the origin and insertion of all fins clearly visible. The final morphometric analysis used to map the phylogeny into the morphospace included only those species for which combined-evidence phylogenetic results were available, a total of 24 species.
Thirteen landmarks were chosen to demonstrate major features of body shape exclusive of fin shapes, which could not be reliably measured in the available images or specimens. Absolute sizes of imaged specimens were often not available and were not needed for this type of analysis. Landmarks were digitized in ImageJ version 2.0.0 (Rasband, 2016) using the Point Picker plugin (Thévenaz, 2016). Morphometric analysis was conducted in MorphoJ version 1.06d (Klingenberg, 2011). Landmarks were subjected to Procrustes fit aligned by principal axes. A covariance matrix was generated from the Procrustes coordinates. Procrustes coordinates were subjected to principal component analysis (PCA). Species were classified by family following Tyler and Santini (2005). Shape changes corresponding to each of the first four principal components of the PCA were analyzed using wireframe deformation diagrams. The combined-evidence phylogeny was mapped into the PCA morphospace to create a phylomorphospace.
RESULTS
Phylogeny based on DNA sequences.—The maximum likelihood (ML) tree from each of the two optimizations [log(ML) = –43651.8 and –43652.1] recovered the same branching pattern (Fig. 2) and reflected some but not all of the relationships recovered in the Bayesian (BI) analysis (Fig. 3; log of model likelihood = –43806.95). Notably, the families Cyttidae, Grammicolepididae, Oreosomatidae, and Zeidae were recovered as monophyletic (ML: 100% bootstrap support; BI: 1.00 posterior probability), and with memberships consistent with those identified by Tyler et al. (2003) and Tyler and Santini (2005). With respect to Oreosomatidae, in both the ML and BI analyses, Oreosoma is sister to Allocytus + (Pseudocyttus + Neocyttus). This is contrary to the morphological results of both Tyler et al. (2003) and Tyler and Santini (2005), who found Pseudocyttus to be sister to the others, although they disagreed about relationships among the other three genera.
Fig. 2.
Maximum likelihood (ML) phylogeny of the Zeiformes as reconstructed by Garli v2.0 (Zwickl, 2006), using sequence data for the eight molecular loci of Table 1, under the substitution models given in Table 2. Support values at nodes are bootstrap percentages. For details of the outgroup relationships see Figure 1S (see Data Accessibility). Asterisk (*) indicates sample originally cataloged as Cyttomimus affinis. Numbers after scientific names correspond to code numbers in Table 1.
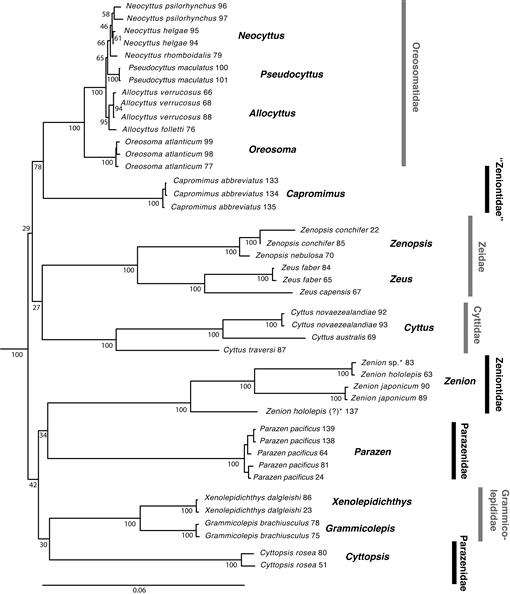
Fig. 3.
Bayesian inference (BI) phylogeny of the Zeiformes as reconstructed by MrBayes v.3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003), using sequence data for the eight molecular loci of Table 1, under the substitution models given in Table 2. Support values at nodes are posterior probabilities. For details of the outgroup relationships see Figure 2S (see Data Accessibility). Asterisk (*) indicates sample originally cataloged as Cyttomimus affinis. Numbers after scientific names correspond to code numbers in Table 1.

Zeniontidae (Capromimus and Zenion in our analyses), on the other hand, were polyphyletic in both ML and BI results, with Capromimus removed from Zeniontidae and placed with strong support (ML: 78%, BI: 1.00) as the sister group of Oreosomatidae. The position of Cyttomimus, the third member of the Zeniontidae according to Tyler et al. (2003), could not be tested with molecular data because the tissue obtained for this genus was found to represent a species of Zenion. Concerning the Parazenidae (Cyttopsis, Parazen, and Stethopristes as per Tyler et al., 2003), ML and BI results differed. The two genera included in our molecular data (Cyttopsis, Parazen) were united in the Bayesian analysis with a posterior probability of 0.61 (Fig. 3), but in the ML results, Cyttopsis was instead sister to Grammicolepididae, albeit with very weak (30%) bootstrap support (Fig. 2). Although support is again low, there is also phylogenetic signal in both ML and BI analyses (Figs. 2, 3) for a group containing Parazen, Cyttopsis, Zenion, and Grammicolepididae (42% and 0.93, respectively).
Concerning the early branching of extant taxa, our BI analysis of molecular data places Zeidae with weak support as sister to all other Zeiformes, in contrast to Tyler et al. (2003), who recovered Cyttidae as sister to all other zeiforms. The present ML results divide the order into two larger clades also with weak support, but different memberships than the two larger clades suggested by Tyler and Santini (2005).
When the revised outgroups were replaced by six genera used as outgroups by Tyler et al. (2003) to evaluate the effect of outgroup choice, both the ML and BI analyses (not shown) recovered a monophyletic Zeiformes with very strong support (MI: 100%, BI: 1.00). A non-monophyletic Zeniontidae, with Capromimus sister to Oreosomatidae, was once again recovered with strong support (ML: 98%, BI: 1.00). Parazenidae were also recovered as non-monophyletic in the ML analysis, with Cyttopsis forming the sister to Grammicolepididae (34%), congruent with the ML results from revised outgroups. Parazen + Zenion was recovered in both ML (51%) and BI (0.88) analyses, congruent with our ML (revised outgroups) analysis and that of Tyler and Santini (2005). Although Grammicolepididae were recovered as sister to all other zeiforms in the BI analysis with weak support (0.53), Zeidae were recovered as sister to all others in the ML analysis (100%), congruent with the BI analysis with revised outgroups, but incongruent with both Tyler et al. (2003) and Tyler and Santini (2005).
Phylogeny based on morphology.—Based on parsimony, a single shortest tree of 319 steps was recovered and rooted on Polymixia (Fig. 4). Of the 105 characters analyzed, two are invariant and four are uninformative, leaving 99 informative characters (many of them multi-state). Tree statistics are: CI: 0.567; RI: 0.735; RC: 0.417; HI: 0.433. Not surprisingly, zeiforms were recovered as monophyletic with 99% bootstrap support and decay value of eight, consistent with the findings of Tyler et al. (2003), Tyler and Santini (2005), Grande et al. (2013), and our molecular analyses herein. The following internal zeiform relationships were recovered in the present morphological parsimony analysis: Zeidae + [Macrurocyttus + (Grammicolepididae + ((Cyttidae + (Parazenidae + Zeniontidae)) + Oreosomatidae))]. All zeiform families except Grammicolepididae (Xenolepidichthys + Grammicolepis + Macrurocyttus, as per Tyler et al., 2003) were recovered as monophyletic, but with varying degrees of bootstrap/decay support. Zeidae, Oreosomatidae, and Cyttidae exhibit the strongest bootstrap and decay support (99%/≥10, 70%/3, and 92%/5, respectively). Zeidae were recovered as the earliest diverging zeiform clade, but with weak bootstrap and decay support (17%/2) for the clade of all other Zeiformes. Grammicolepididae, consisting only of Xenolepidichthys + Grammicolepis in these results, were highly supported (100%/≥10). Within Parazenidae, Cyttopsis and Stethopristes were recovered as sisters with strong support (97%/5), although Parazen was included in the family as sister to Cyttopsis + Stethopristes only with weak support (36%/2). This tenuous relationship is consistent with the results of our molecular analysis and not very different from those of Tyler and Santini (2005) and Grande et al. (2013), both studies finding Parazenidae to be non-monophyletic. Zeniontidae were recovered as monophyletic but not strongly supported in this analysis (34%/3), a finding somewhat different from that of our molecular analyses, which recovered Capromimus sister to Oreosomatidae with 78% bootstrap support.
Fig. 4.
Maximum parsimony (MP) phylogeny of Zeiformes based on morphological data analyzed in PAUP v.4b10 (Swofford, 2003). The data (Table 3) are a modified version of those used by Tyler and Santini (2005) but with revised outgroups reflecting zeiform membership in the Paracanthopterygii (e.g., Grande et al., 2013), and with character deletions, additions, and edits described in the text. The characters are listed in Appendix 1. This analysis is based on 27 terminal taxa and 105 characters, and resulted in a single shortest tree of 319 steps with CI 0.567. Support values at nodes are 1000-replicate bootstrap percentages/decay (Bremer) values. Character-state optimization was in MacClade 4.08 (Maddison and Maddison, 2005) using the ACCTRAN option as used also by Tyler et al. (2003) and Tyler and Santini (2005). Thumbnail drawings of representative species are original artwork by Michael Hanson.
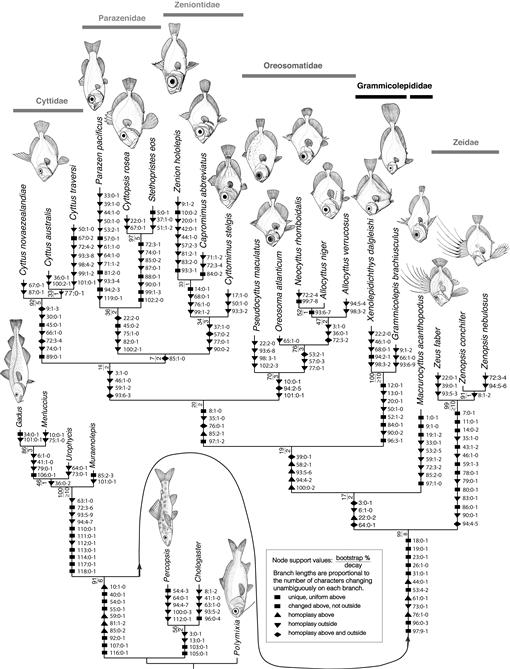
Fig. 5.
Comparison of branching patterns of morphological trees under maximum parsimony (MP) from three studies. Each family recognized by Tyler et al. (2003) is a different color. (A) Results from Tyler et al. (2003). (B) Results from Tyler and Santini (2005). (C) Results from the morphological analysis of the present study. Black lines in C indicate relationships with strong support; gray lines indicate weaker support. Note that the new tree resembles the previous trees but with the root moved to a position near Zeidae.
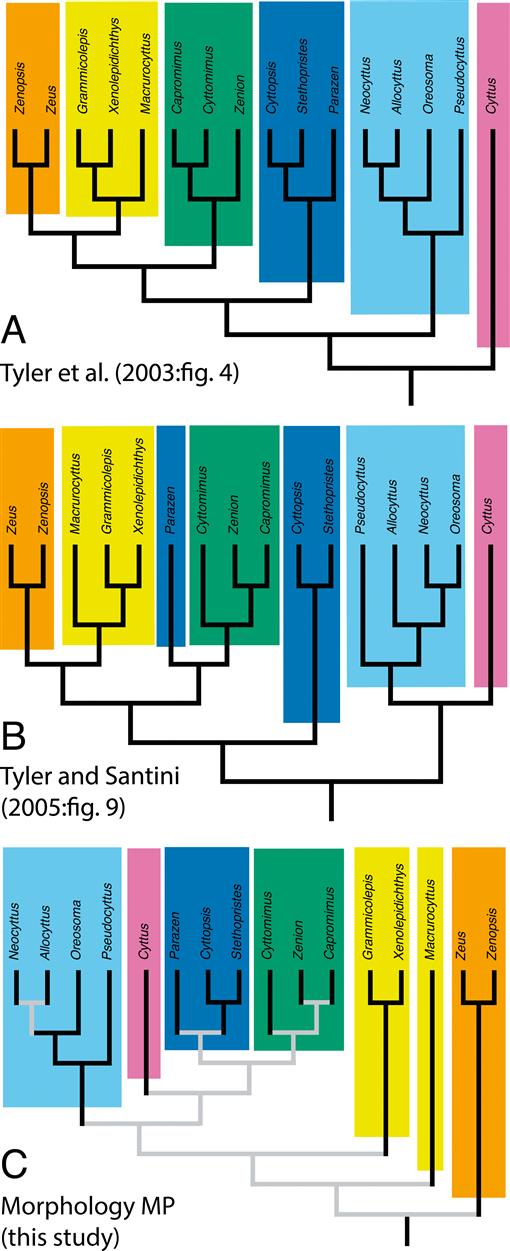
Results of our morphological parsimony analysis both support and, especially in the placement of the root, conflict with the findings of previous authors (Fig. 5). For example, Tyler et al. (2003) recovered a fully resolved phylogeny based on morphological characters, although with different outgroups, and with the following relationships: Cyttidae + {Oreosomatidae + [Parazenidae + (Zeniontidae + (Grammicolepididae + Zeidae))]}. In their results, Cyttidae diverged earliest and Zeidae + Grammicolepididae were considered highly derived. The differences between their results and those of the present study are attributable largely to a different root placement, near Zeidae in the present study but near Cyttidae in Tyler et al. (2003). In Tyler and Santini (2005), zeiform interrelationships were reexamined but this time with the inclusion of several fossil taxa (notably †Cretazeus and the two fossils from Denmark). This time Parazenidae were not recovered as monophyletic, and Parazen grouped with Zeniontidae. In the latter analysis, the fossil zeiforms from Denmark were successively sister to all other Zeiformes, which were divided into two groups as follows: [Oreosomatidae + Cyttidae] and [Parazenidae (Zeniontidae (Grammicolepididae + Zeidae))], where their clade “Parazenidae” excluded Parazen, which they placed in Zeniontidae. The authors argued that the inclusion of fossils with a preponderance of missing data might have affected their results. Their results also differ notably from those of the present study in the placement of the root, near Zeidae in our results versus near Cyttidae + Oreosomatidae. The similarity between our results and those of the two earlier studies, except for root placement resulting from revised outgroups, is evidence of a strong phylogenetic signal that originated with Tyler et al. (2003), persisted through addition of data on fossils by Tyler and Santini (2005), and has been maintained through revisions and additions to the data in the present study.
Phylogeny based on combined data.—Like the separate molecular and morphological analyses, the combined (total evidence) morphological plus molecular analysis (Figs. 6, 7) also supports the monophyly of Zeiformes and, with the root near Polymixiiformes, the following relationships: Percopsiformes (Zeiformes [Stylephoriformes + Gadiformes]). These relationships are the same as those from our separate molecular and morphological analyses (Figs. 2–4, S1, S2; see Data Accessibility).
Fig. 6.
Combined (total-evidence) molecular and morphological phylogeny of the Paracanthopterygii based on Bayesian inference (BI) using MrBayes v.3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). See text for detailed methods and assumptions. Zeiform taxa with an asterisk (*) are those with morphological data only. Support values at nodes are posterior probabilities. For details within Zeiformes see Figure 7. Drawings of representative species by Michael Hanson.
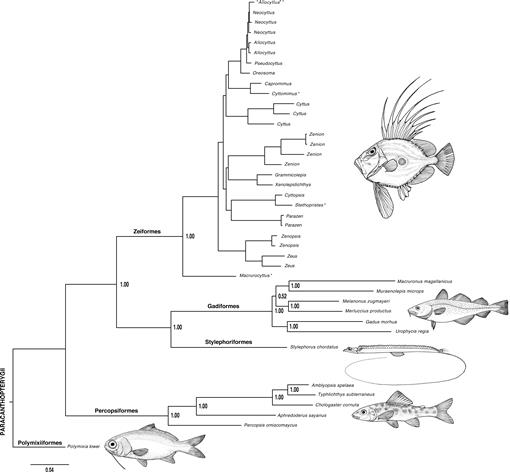
Fig. 7.
Combined (total-evidence) molecular and morphological phylogeny of the Zeiformes based on Bayesian inference (BI) using MrBayes v.3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). See Figure 6 for details of outgroup relationships, and see text for detailed methods and assumptions. The combined maximum likelihood (ML) analysis using Garli v2.0 (Zwickl, 2006) produced almost identical topology and very similar relative branch lengths. Taxa with asterisks (*) are of questionable or revised identification. Taxa with two asterisks (**) are those with morphological data only. Support values at nodes are from both analyses, with BI posterior probabilities above ML bootstrap percentages. Numbers after scientific names correspond to code numbers in Table 1. Thumbnail drawings of representative species by Michael Hanson.
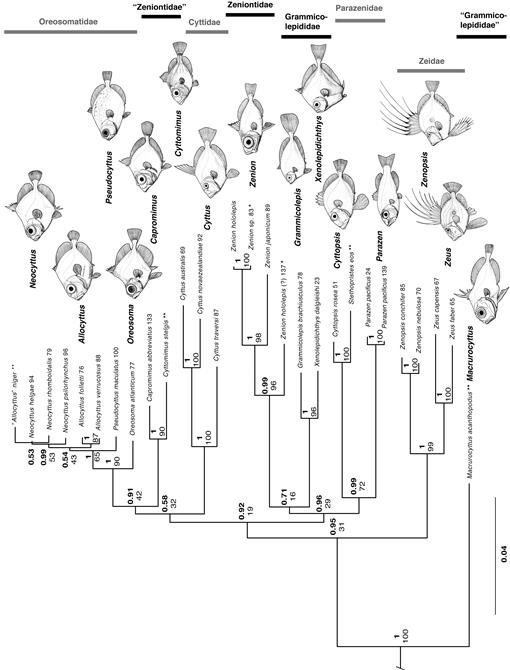
The BI (log of model likelihood = –39031.86) and ML (ln[likelihood] = –38831.94) combined hypotheses shared several other features with the DNA-only analyses, namely the compositions of families for genera for which DNA data were available and the polyphyly of Zeniontidae (Fig. 8). Because the branching topologies of the ML and BI trees were nearly identical (Fig. 8C, D), both sets of nodal support values are shown in Figure 7, which shows the BI tree. In general, nodal support was greater for the BI combined analysis than for the ML combined analysis.
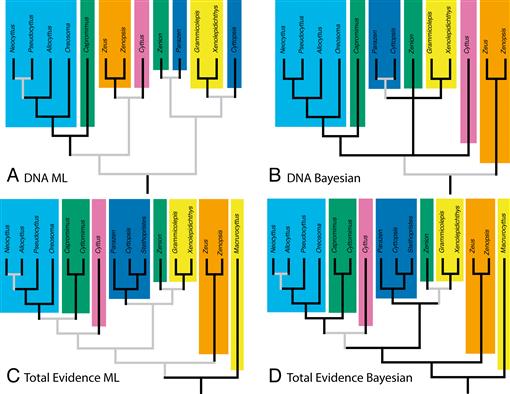
Fig. 8.
Comparison of the branching patterns of molecular and combined-data trees produced with different methods. Each family recognized by Tyler et al. (2003) is a different color. Black lines indicate relationships with strong support; gray lines indicate weaker support. (A) Results from maximum likelihood analysis of molecular data. (B) Results from Bayesian inference analysis of molecular data. (C) Results from maximum likelihood analysis of combined morphological and molecular data, including three genera with morphological data only. (D) Results from Bayesian inference analysis of combined morphological and molecular data, including three genera with morphological data only.
Relationships recovered within Zeiformes are: Macrurocyttus (Zeidae ((Parazenidae (Zenion + Grammicolepididae)) (Cyttidae ((Capromimus + Cyttomimus) Oreosomatidae)))), where Grammicolepididae does not include Macrurocyttus. Two previously recognized families (Tyler et al., 2003) are polyphyletic in these results: Grammicolepididae and Zeniontidae (Figs. 7, 8C, D).
Among the four taxa represented by morphological data only, Allocyttus niger was recovered within Oreosomatidae as expected, but in a closer relationship to Neocyttus spp. than to other Allocyttus spp. (Fig. 7). The low support values make this relationship very tentative.
Cyttomimus, also represented only by morphological data (the tissue obtained for this genus was shown by our results to belong in Zenion), was recovered with strong support as close to Capromimus, with those two genera as the sister group of Oreosomatidae (Figs. 7, 8). Both genera were in Zeniontidae in Tyler et al. (2003) and Tyler and Santini (2005) and in our morphological analysis (Figs. 4, 5). However, the Zeniontidae are polyphyletic in both the ML and BI molecular analyses (Figs. 2, 3) and in both the ML and BI combined analyses herein (Figs. 7, 8).
Stethopristes was recovered on morphological data only as sister to Cyttopsis (Figs. 7, 8), with those two being sister to Parazen in these combined results. This supports monophyly of the Parazenidae as proposed by Tyler et al. (2003) and supported also by our morphological analysis (Fig. 4), although Tyler and Santini (2005) later recovered Parazen as sister to Zeniontidae (Fig. 5).
The rare Macrurocyttus acanthopodus was also represented by morphological data only in the present study. Represented by probable larval specimens, it was placed in Grammicolepididae by Tyler et al. (2003) and Tyler and Santini (2005), but in our morphological analysis (Figs. 4, 5) it was recovered as separate from Grammicolepididae and sister to all zeiforms except Zeidae. In the combined analysis of the present study, it was recovered as sister to all zeiforms including Zeidae (Figs. 7, 8), with a posterior probability of 0.95 in the BI analysis but only 31% bootstrap in ML.
Morphometric results.—Principal components analysis of Procrustes coordinates from 13 body-shape landmarks (Fig. 9A) of 24 zeiform species produced principal components of which the first three explain more than 75% of the total variance (PC1 36.5%, PC2 22.2%, and PC3 16.4%; Fig. 9B–D). The wireframe for PC1 (Fig. 9B) and the morphospace in Figure 9E illustrate that the first component corresponds to differences in body shape from the average form with deeper body, smaller and higher orbit, and longer jaws toward forms such as Parazen and Zenion with shallower body, relatively larger eye placed more laterally, and shorter jaws (Fig. 9B, E). In the opposite direction, low values of PC1 correspond to fishes such as Cyttus traversi, Oreosoma, and Cyttopsis with deep bodies, eyes placed more dorsally, and longer jaws. The PC2 wireframe (Fig. 9C) shows changes from the average toward a relatively smaller orbit, more oblique jaws, and more anteriorly placed paired fins. As shown also in Figure 9E, PC2 separates taxa such as Macrurocyttus, Cyttopsis, Allocyttus verrucosus, and Neocyttus rhomboidalis with larger heads and more terminal mouths from genera such as Zeidae and two of three genera of Grammicolepididae with smaller heads and more oblique mouths (Fig. 9E). PC3 (Fig. 9D) separates taxa such as Xenolepidichthys, Grammicolepis, and Neocyttus helgae with shorter jaws and more lateral orbit from taxa such as Zeus, Zenopsis, Stethopristes, and Cyttopsis with longer jaws and smaller, higher orbit.
Fig. 9.
Morphometric analysis of 24 zeiform species for which combined molecular and morphological phylogenetic results were obtained as in Figure 7. (A) Thirteen landmarks used in the morphometric analysis shown on an image of Zenopsis nebulosa (CSIRO). (B–D) Wireframe diagrams in which the red wireframe represents an average form while the blue outline represents change in the direction of the principal component. (E) Phylomorphospace diagram showing variation in the morphospace defined by the first two principal components, with the combined phylogeny of Figure 7 mapped into the morphospace. Thin blue lines and thick black lines represent convergent lineages. Note that most of the major lineages arose from common ancestors with near-average morphologies (blue ellipse). Thumbnail drawings by Michael Hanson.
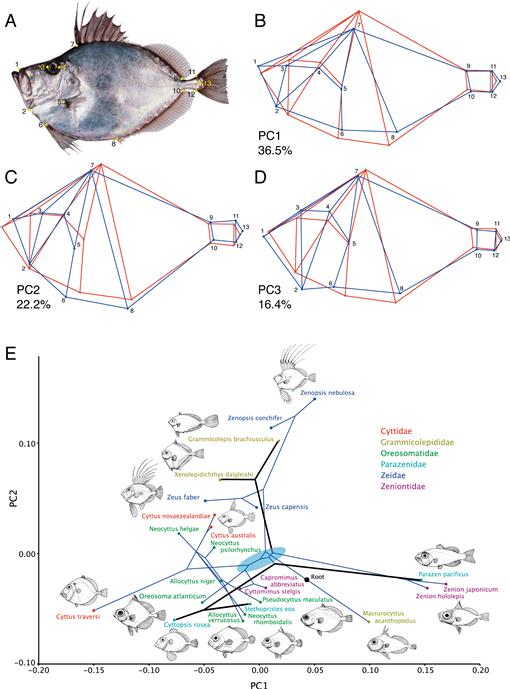
These results emphasize the strong differences in body shape contained within the families Grammicolepididae, Parazenidae, and Zeniontidae as previously understood. All three families have been recovered as polyphyletic in recent analyses, Parazenidae by Tyler and Santini (2005), and Grammicolepididae and Zeniontidae in our combined-data results (Fig. 7). Parazen scored high on PC1, whereas Stethopristes and Cyttopsis, the other two parazenid genera, scored low. On PC2, scores for the three genera were more similar, but on PC3 and PC4 they were again widely separated. The three monotypic genera of Grammicolepididae were separated by PC1 and PC2 such that Macrurocyttus scored high on PC1 and low on PC2, whereas Grammicolepis and Xenolepidichthys scored at the opposite extreme for PC1 and PC2 (Fig. 9E). Similarly, the family Zeniontidae contains disparate body forms, with Zenion at one extreme and Capromimus and Cyttomimus having closer to average scores on the first two principal components.
The phylomorphospace analysis (combined-evidence tree mapped into PCA morphospace; Fig. 9E) suggests that the common ancestor of extant zeiforms was one with a near-average body form (blue ellipse in Fig. 9E). The small size and probable late larval body form of the imaged specimen of Macrurocyttus demand caution in interpreting its body shape as primitive for the order. The concentration of lineage branching near the center (0, 0) of the morphospace (blue ellipse), with body forms similar to those of Capromimus and Cyttomimus, is consistent with the short branches near the base of the zeiform radiation in the molecular phylogenetic trees (Figs. 2, 3). Strongly divergent lineages radiate in at least three directions of change and denote marked convergence (Fig. 9E). Change toward shallow bodies, large eyes, and strongly oblique mouths characterize convergence in two lineages (Parazen, indicated by a thick black line, and Zenion, indicated by thin blue lines). Change toward deep bodies with less oblique mouths occurred convergently in the lineages leading to Cyttidae + Oreosomatidae (thin blue lines) and in the ancestry of Cyttopsis and Stethopristes (thick black lines). Change toward relatively smaller heads, smaller and higher eyes, and more oblique jaws occurred in Zeidae (Zeus, Zenopsis; thin blue lines) and separately in the ancestry of two of three genera of Grammicolepididae (Grammicolepis, Xenolepidichthys; thick black lines).
DISCUSSION
This study represents an integrative approach that includes morphology, molecules, and morphometrics and was able to resolve many outstanding issues in the evolution of Zeiformes. However, there remain unanswered questions about the relationships of certain genera and species. In particular, molecular evidence for the placement of Allocyttus niger, Cyttomimus, Stethopristes, and Macrurocyttus is still wanting because tissues suitable for molecular analysis are needed. The relationships of Macrurocyttus are especially important given its earliest diverging position in our combined tree, its early diverging position (sister to all except Zeidae) in our morphological tree, and the rarity or absence of juvenile or adult specimens in collections. If corroborated by molecular evidence, Macrurocyttus could once again be recognized in its own family, Macrurocyttidae Myers, 1960, as per Heemstra (1999). Based on strong molecular evidence, Capromimus does not belong in Zeniontidae and is sister to Oreosomatidae. Cyttomimus is its closest relative based on our combined analysis. If these results are supported in the future by additional morphological and molecular data, these two genera could be recognized as a distinct subfamily of Oreosomatidae.
Seeing Zeiformes from a different perspective (i.e., as paracanthopterygians) requires us to revise our ideas about their origin and diversification, including the timing of their radiation, their ancestral body plan, the multiple examples of convergence, the speed of their radiation, and possible adaptive explanations. Concerning timing, the relevant facts include the ages of older fossils as well as those of their close relatives in Paracanthopterygii. Ages cited here are based on stratigraphic occurrences, with absolute ages from Cohen et al. (2016). The oldest fossil zeiforms are discussed above and include †Cretazeus of Campanian/Maastrichtian age (∼72 Ma), the two small fossils from the late Paleocene/early Eocene (∼56 Ma), and †Bajaichthys from the early Eocene (late Ypresian, ∼50 Ma). The most significant of these is †Cretazeus because, although it is much older than the other fossils, it is suggested to be within the crown Zeiformes in “Parazenidae” (Tyler and Santini, 2005; Davesne et al., 2017; however, Parazen was not recovered by either study in the same clade).
To assess the relationships of the four fossil zeiforms, we added each one to a separate analysis, with the phylogeny constrained to the topology of our combined analysis (Figs. 6, 7), and searched for the placement of each one that minimally increased tree length under parsimony. For all four fossils, placement as sister to all zeiforms except Zeidae and Macrurocyttus caused minimal length increase. For two, a second placement was equally parsimonious: “†Archaeozeus” as sister to Cyttidae + Oreosomatidae, and †Cretazeus as sister to Parazenidae. All of these placements differ from those proposed by Tyler and Santini (2005) and Davesne et al. (2017), who suggested †Cretazeus to be sister to Cyttopsis + Stethopristes and the other three to be stem zeiforms. Regardless of which placements are correct, the initial radiation of extant zeiform lineages must have pre-dated the age of the oldest fossil, †Cretazeus, near the Campanian–Maastrichtian boundary.
The ages of the oldest fossils of all paracanthopterygians further suggest that most of the lineages within Paracanthopterygii (Fig. 6) evolved during the Late Cretaceous (100–66 Ma). These include the gadiforms (Kriwet and Hecht, 2008), the oldest fossils of which are of Paleocene age. There are no known fossils of Stylephoriformes. The oldest described fossil percopsiforms based on articulated skeletons are the early Paleocene †Mcconichthys Grande, 1988 (family †Mcconichthyidae) from Montana (∼65 Ma) and two late Paleocene (∼58 Ma) genera of Percopsidae from Alberta (Murray and Wilson, 1999). Disarticulated dentaries from the Maastrichtian (∼70 Ma) of Montana (Brinkman et al., 2014) and the mid-Campanian (∼77 Ma) of Alberta (Neuman and Brinkman, 2005) were recently also assigned to the Percopsiformes, the oldest records to date for this order. The extinct paracanthopterygian order †Sphenocephaliformes includes two genera and four species, two species in the Campanian (∼77 Ma) genus †Sphenocephalus and two in the very early Cenomanian (∼100 Ma) genus †Xenyllion (e.g., Newbrey et al., 2013). The earliest diverging clade in the Paracanthopterygii is the Polymixiiformes (Grande et al., 2013; Chen et al., 2014), appearing in the fossil record in the Cenomanian (∼95 Ma) and represented soon after by a number of Cretaceous genera (e.g., Davesne et al., 2016; Nelson et al., 2016). We conclude that the rapid radiation of the major extant zeiform lineages occurred prior to the Campanian, but likely not earlier than the Cenomanian, since the earliest Cenomanian is the age of the oldest known fossil acanthomorphs.
Concerning the likely ancestral body plan of zeiforms, the revised combined-data phylogeny has Macrurocyttus as the earliest diverging lineage, but taking into account the likely larval life-history stages of known specimens, we do not consider its general body form as representative of the zeiform ancestor. The phylomorphospace analysis suggests that the early radiation of zeiforms involved several initial lineage splits of fishes with near-average morphometric proportions (moderately deep body and long jaws), similar to the body forms of the genera Capromimus and Cyttomimus.
Multiple examples of convergence are illustrated by radiations from this average morphotype. Two spokes of the radiations led convergently to two genera (Parazen, Zenion) with shallow bodies, large, more laterally placed eyes, and smaller, more oblique jaws. Two more spokes led convergently to forms with relatively deep bodies, smaller orbits, smaller heads, and more oblique jaws (Grammicolepis + Xenolepidichthys, Zeus + Zenopsis). A third set of spokes led to forms (Cyttidae, Oreosomatidae, Cyttopsis) with deeper bodies, high orbits, and less oblique jaws.
Results from this study exhibit a pattern of divergence among the major lineages of extant zeiform fishes suggestive of a relatively rapid initial diversification, perhaps similar to an early-burst adaptive radiation such as that proposed by Martin and Wainwright (2013). Although a few zeiform genera were difficult to place, the significant morphological disparity seen among families renders most of the groups easily recognizable, yet this disparity in morphological characters (Figs. 4, 9) is not reflected in major sequence changes within the studied molecular loci, as evidenced by the short branch lengths (Figs. 2, 3) leading to all of the major groups of crown zeiforms. A similar phenomenon may also characterize radiation within the most taxonomically diverse zeiform family, the Oreosomatidae. Significant morphological differences among oreosomatid genera (Figs. 4, 9E) are a contrast to the short branch lengths in the molecular results (Figs. 2, 3).
The star-shaped pattern of diversification in the phylomorphospace (Fig. 9E) is also consistent with the idea that there were multiple fitness peaks in an adaptive landscape (Martin and Wainwright, 2013) during the early evolution of zeiforms, which could have driven strong divergence from the ancestral form and multiple convergences toward disparate body forms.
MATERIAL EXAMINED
Institutional abbreviations follow Sabaj (2016). For a more complete list of outgroups examined for morphology, see Grande et al. (2013). Key: alcohol, preserved in ethanol; CS, cleared and stained for bone and cartilage; x-ray, radiograph examined for morphological characters; image, photos and radiographs digitized for morphometrics.
Gadiformes
Bregmacerotidae.—Bregmaceros cantori: 1 spec., 49.5 mm SL: KU 30244 (CS).
Bregmaceros sp.: 5 spec., 68.0–76.6 mm SL: USNM 398649 (alcohol, CS), USNM 398650 (alcohol).
Gadidae.—Gadinae: Gadus macrocephalus: 2 spec., 120.5–164 mm SL: KU 15063 (CS), LACM 33868 (alcohol).
Gadus morhua: 2 spec., 12.1–103.8 mm SL: ROM 48371 (alcohol), ROM 62449 (CS).
Phycinae: Phycis blennoides: 3 spec., 120.2–128.1 mm SL: USNM 232482 (alcohol, CS).
Phycis chesteri: 2 spec., 130.5–190.5 mm SL: LACM 56741 (CS).
Phycis phycis: 1 spec., 80.0 mm SL: FMNH 69332 (CS).
Urophycis cirrata: 1 spec., 158.5 mm SL: LACM 56745 (CS).
Urophycis earllii: 1 spec., 163.4 mm SL: LACM 56750 (CS).
Urophycis floridana: 3 spec., 109.5–117.4 mm SL: FMNH 51025 (alcohol, CS).
Urophycis sp.: 42 spec., 5–18 mm SL: MCZ 85872, 97634 (alcohol, CS).
Macruronidae.—Macruronus novaezelandiae, 1 spec., 437.8 mm SL: CAS 213332 (alcohol).
Macruronus sp.: 1 spec., 135.0 mm SL: LACM 56759 (CS).
Merlucciidae.—Merluccius albidus: 4 spec., 120.5–151.8 mm SL: FMNH 69318 (alcohol, CS).
Merluccius gayi: 1 spec., 173.1 mm SL: KU 14653 (alcohol).
Merluccius hernandezi: 10 spec., 40–60 mm SL: LACM 8269 (alcohol, CS).
Merluccius productus: 15 spec., 15–145.0 mm SL: LACM 56764 (alcohol, CS), LACM 6700-8 (alcohol, CS).
Percopsiformes
Amblyopsidae.—Amblyopsis spelaea: 4 spec., 60.0–74.2 mm SL: CAS 78143 (alcohol, dissected CS), USNM 44435 (alcohol).
Chologaster cornuta: 4 spec., 28.9–39.1 mm SL: KU 8874 (CS), USNM 237005 (alcohol).
Typhlichthys subterraneus: 1 spec., 40.2 mm SL: USNM 36806 (alcohol).
Aphredoderidae.—Aphredoderus sayanus: 9 spec., 34.5–85.1 mm SL: FMNH 78533 (CS), KU 2412 (CS), KU 5032 (alcohol, CS), KU 33610 (alcohol, CS), USNM 84051 (alcohol), USNM 396352 (alcohol).
Percopsidae.—Percopsis omiscomaycus: 31 spec., 35.6–115.3 mm SL: FMNH 63444 (CS), FMNH 63459 (alcohol, CS), FMNH 86990 (alcohol, CS), KU 3432 (alcohol, CS), KU 7949 (alcohol), KU 10476 (alcohol, CS).
Percopsis transmontana: 23 spec., 49.10–63.0 mm SL: UAMZ F402 (alcohol, CS), USNM 366393 (alcohol, CS).
Zeiformes
Cyttidae.—Cyttus australis: 3 spec.: LACM 42620, 99.6 mm SL (alcohol, x-ray), USNM 177084 (adult, x-ray, not measured); USNM RAD 107357 (image).
Cyttus novaezealandiae: 1 spec.: CSIRO Australian National Fish Collection via Fishes of Australia (image).
Cyttus traversi: 1 spec.: USNM 308020, 105.8 mm SL (alcohol, x-ray, image).
Grammicolepididae.—Grammicolepis brachiusculus: 3 spec., 175–200 mm SL: FMNH 74298 (CS), MCZ 44910 (x-ray, not measured), USNM 227936 (image).
Grammicolepis latiusculus: 1 spec., 25 mm SL: MCZ 57868 (alcohol).
Macrurocyttus acanthopodus: 3 spec.: NMV A25383-2 (image), USNM 367331 (2 spec., adult, x-ray).
Xenolepidichthys dalgleishi: 36 spec., 7–100.4 mm SL: MCZ 162020 (alcohol), MCZ 57867, 57869, 57871, 57872, 84958, 155446 (8–20 mm SL, images), USNM 320016 (CS), USNM 377985 (alcohol, CS, image), USNM 398654 (alcohol, CS).
Oreosomatidae.—Allocyttus niger: AM I.33319-001 (image), MNHN IC-2000-1360 (x-ray, image).
Allocyttus verrucosus: 1 spec., 139 mm SL: LACM 44752-2 (alcohol, x-ray).
Neocyttus helgae: 2 spec.: MNHN 1998-0726 (x-ray), Fishbase D Devitt id R Bañon Diaz, Nehel_u8 (image).
Neocyttus psilorhynchus: CSIRO Australian National Fish Collection via Fishes of Australia (image).
Neocyttus rhomboidalis: AM I.20097-001 (image), CSIRO H6054-02/NORFANZ (image).
Oreosoma atlanticum: 3 spec., 112.85–127.6 mm SL: KU 33415 (alcohol, CS, image), USNM 385874 (x-ray, juvenile, not measured).
Pseudocyttus maculatus: MNHN I-2000-1361 (image).
Parazenidae.—Cyttopsis cypho: 1 spec., USNM RAD 93140 (adult, x-ray).
Cyttopsis rosea: 10 spec., 18.0–97.6 mm SL: FMNH 67091 (CS), MCZ 85100, 85101 (alcohol), USNM 50562 (holotype of Cyttopsis itea), USNM 186029, (x-ray, adult, not measured), USNM 377980 (alcohol, CS).
Parazen pacificus: 7 spec., 71.30–110.7 mm SL: ASIZP 0066019 (x-ray, adult not measured), FMNH 67158 (alcohol, CS), MCZ 40029 (image), USNM 364277 (alcohol, CS).
Stethopristes eos: 6 spec., 45–105.2 mm SL: USNM 226570 (CS), USNM RAD 51626 (x-ray, image), USNM RAD 51685 (x-ray, adult, not measured).
Zeidae.—Zenopsis conchifer: 13 spec., 70.3–109.0 mm SL: KU 26983, 26985 (alcohol, CS, image), USNM 372241 (alcohol, CS).
Zenopsis nebulosa: 1 spec.: ASIZP 0066135 (adult, x-ray), CSIRO via Fishes of Australia (image).
Zenopsis ocellatus: 3 spec., 68.9–73.0 mm SL: FMNH 67179 (CS), USNM 159819 (alcohol, CS, image).
Zeus faber: 8 spec., 49.8–110.8 mm SL: USNM 307842 (CS), USNM 325986 (alcohol, CS, image).
Zeniontidae.—Capromimus abbreviatus: 1 spec., 60.4 mm SL: LACM 11490-1 (CS).
Cyttomimus affinis: ASIZ P.0058509 (x-ray, adult not measured), NMNS Taiwan F0079 (image).
Zenion hololepis: 19 spec., 48.3–86.8 mm SL: USNM 377986 (alcohol, CS, image), USNM 380010 (alcohol, CS).
Zenion leptolepis: MNHN RAD 1996-1479 (x-ray, adult, not measured, image).
Zenion sp.: 5 spec.: MNHN IC-2000-1458 (x-ray).
DATA ACCESSIBILITY
DNA sequences are deposited in GenBank under accession numbers given in Table 1. Matrices and supplemental figures are available at http://www.copeiajournal.org/cg-17-594.
ACKNOWLEDGMENTS
We thank the following people and institutions for specimen or tissue loans: K. Hartel (MCZ), C. McMahan (FMNH), C. Thacker (LACM), J. Bruner and A. Murray (UAMZ), R. Winterbottom and M. Burridge (ROM), E. Hilton and S. Huber (VIMS), M. Lisher and M. Anderson (SAIAB), A. Graham (CSIRO–Hobart), L. Page and R. Robins (FLMNH), P. McMillan (NIWA–Taihoro Nukurangi), D. Bray (NMV), M. McGrouther (AMS), P.-L. Lin (ASIZP), R. Mayden (SLUC), B. Kuhajda (UAIC), H. Walker, Jr. (SIO), L. Smith and A. Bentley (KU), L. Parenti, C. Baldwin, and J. Williams (USNM), G. Ortí (GWU). We thank S. Reddy (Loyola University Chicago) for the use of her molecular lab to carry out the molecular protocols for this project. Our sincerest thanks to J. Tyler for his insight and discussions about zeiform fishes. We thank D. Davesne and J. Tyler for their thoughtful comments. We thank C. Li (Shanghai Ocean University), S. Reddy, and L. Smith for discussion regarding analyses. Many thanks go to J. Young and J. Peters for their help with the morphometric analysis. Special thanks go to M. Hansen for his artistic expertise in executing original drawings. This research was funded in part by The National Science Foundation (NSF DEB-0732589) to Grande, an American Museum Lerner-Gray Grant to Scarpitta, and by NSERC (Canada) Discovery Grant 9180 to Wilson.
LITERATURE CITED
Appendices
APPENDIX 1
Complete list of characters used in the morphological study
Characters 1–103 are from Tyler and Santini (2005) and form the foundation of our character matrix. Some of these characters overlapped with Grande et al. (2013) or Borden et al. (2013). Those character numbers are given in parentheses when applicable. Characters 104–113 are from Grande et al. (2013), and characters 114–119 are from Borden et al. (2013). Characters 21, 40, 46, 53, 58, 60, 61, 70–72, 80, 81, 86–88, 92, 94, 100, 101, and 103 were modified from Tyler and Santini (2005).
1. Parietal: present (0); absent (1).
2. Basisphenoid: present as a moderate to long, oblique shaft connecting the parasphenoid and the prootic in front of the posterior myodome (0); present as a short shaft at the front of the roof of the posterior myodome (1); absent (2).
3. Vomer, teeth: present (0); absent (1).
4. Parasphenoid opening into the posterior myodome: absent (0); present (1).
5. Skull, opercles, and lachrymal-infraorbitals, with honeycomb bone sculpturing: absent (0); present (1).
6. Frontal, supraocular serrations: present (0); absent (1).
7. Otolith, shape: moderate to large size, rounded or slightly to deeply indented on one or both sides, or oblong with humps (0); tiny, trilobed (bow-tie shaped) (1).
8. Lachrymal, size/depth: large, deep, height about one to four times in the length (0); moderate, height about five to seven times in the length (1); slender (2); not applicable, when absent (‘–').
9. Infraorbitals, number (well-developed elements exclusive of the lachrymal, dermosphenotic, and of variable rudiments): none (0); three or four (1); five or six (2); seven or eight (3); nine or ten (4); 11 or 12 (5).
10. Infraorbitals, depth of most: relatively slender and tubular (0); deep, with large pores and bridges or open lacunae between the upper and lower edges (1); deep, with serrate vertical supporting flanges (2); not applicable, when infraorbitals absent (‘–').
11. Dermosphenotic: a distinctly separate ossification from the sphenotic, sometimes relatively free from the skull (0); fused or highly consolidated with the sphenotic (1); absent as an identifiable part of the sphenotic (2).
12. Mouth, size: large, alveolar process of the premaxilla equal to or longer than the lateral ethmoid depth (0); small, alveolar process no greater and usually much less than the lateral ethmoid depth (1).
13. Postmaxillary process: present (0); absent (1).
14. Premaxilla, alveolar process: simple (0); ventrally indented to form a pair of blunt lobes (1); deeply bifurcated ventrally (2).
15. Premaxilla, ascending process: reaching to a level in front of the orbit or to about the front of the orbit and the lateral ethmoid, no more than the level of 1/5 into the orbit (0); reaching distinctly behind the lateral ethmoid to about the level of 1/3 into the orbit (1); reaching to about the level of ½ into the orbit (2); reaching to the level of the rear of the orbit (3).
16. Palatine, teeth: present (0); absent (1).
17. Ectopterygoid, teeth: present (0); absent (1).
18. Palatine, articulation with the cranium: the main axis of the palatine is relatively parallel, or only moderately oblique, to the body axis and has a fixed, dual articulation with the lateral ethmoid and the ethmo-vomerine region (0); the palatine is usually orientated distinctly obliquely to the body axis and has a single, pivotal, articulation with the lateral ethmoid, resulting in considerable mobility (1).
19. (3) Metapterygoid, size and articulation: relatively large, length c. 3/4 or more of the length of the quadrate, and articulating with it (0); reduced, length c. ½ or less of the length of the quadrate, and not articulating with it (1); absent (2).
20. Symplectic, ventral flange: absent (0); present (1).
21. Dentary, cartilages (on lateral surface of dentary): absent or unconsolidated (0); two moderate cartilages, each attached anteriorly to the dentary and lying sequentially one behind the other, the first shorter than the second (1); two moderate cartilages, each attached anteriorly to the dentary and lying sequentially one behind the other, of about the same size or the first only slightly shorter than the second (2).
22. Dentary, serrations on the lower border: none (0); a single barb near the symphysis (1); multiple serrations behind the symphysis (2).
23. Gill slit, opening between the fourth and the fifth ceratobranchials: present (0); absent (1).
24. Gills, number: four complete gills (one complete gill of two hemibranchs on each complete gill arch), or eight hemibranchs (0); three and a half gills, or seven hemibranchs, with no hemibranch on the rear of the fourth ceratobranchial (1).
25. Gill rakers, number of series on the branchial arches: four and a half, a series present along the rear of the fourth gill slit (at least dorsally), i.e., along the anterior border of the fifth ceratohyal (0); four, no series along the rear border of the fourth gill slit (1); three and a half, no series along the posterior border of the fourth ceratobranchial and none along the rear border of the fourth gill slit (2).
26. First epibranchial, uncinate process: absent (0); present (1).
27. Interarcual cartilage: absent (0); present (1).
28. Second pharyngobranchial, suspensory shaft: short or absent (0); moderately long (1); long, c. ½ to 2/3 the length of the first pharyngobranchial (2).
29. Third pharyngobranchial, suspensory shaft: short or absent (0); moderately long (1); long, almost as long as the shaft of the second pharyngobranchial (2).
30. First basibranchial, position: the upper surface is level with those of the basihyal and the second basibranchial, at least posteriorly (0); the upper surface is entirely below the level of the dorsal surface of the basihyal and the second basibranchial (1).
31. Fourth upper pharyngeal toothplate: present (0); absent (1).
32. Fifth ceratobranchial: toothed (0); toothless (1).
33. (11) Beryciform foramen: present as a completely enclosed opening (0); a deep groove along the lateral surface of the ceratohyal, often onto the dorsal hypohyal (1); a deep concavity on the dorsal surface of the ceratohyal (2); no lateral groove and no deep dorsal concavity, i.e., no deeper than ventral concavity (3).
34. Branchiostegal rays, placement of the heads of the rear group: over the surface or along the ventral edges of both the ceratohyal and epihyal (0); clustered along the ossified posterior border of the ceratohyal (1).
35. Ceratohyal, notches on the lower border: prominent notches for the heads of some of the branchiostegal rays in the anterior group (0); no prominent notches (1).
36. Ceratohyal–epihyal articulation: exclusively through cartilage (0); through cartilage, but with bony interdigitated articulations in some specimens, especially with increasing specimen size (1); bony interdigitated articulations in all specimens at all sizes (2).
37. Epihyal, depth of the anterior end: equal, or almost equal, to the depth of the adjacent part of the ceratohyal (0); distinctly less deep than the adjacent part of the ceratohyal (1).
38. Urohyal, size: small to moderate, no longer than the ceratohyal (0); large, longer than the ceratohyal (1).
39. First vertebra in the caudal peduncle with a modified neural or haemal spine: second preural centrum, PU2 (0); third preural centrum, PU3 (1).
40. (14) First vertebra, association of the neural arch and spine with the skull: the neural arch and spine are not closely applied to the skull (0); the neural arch and spine are closely applied to the skull, primarily to the exoccipitals (1).
41. Second and subsequent few anterior abdominal vertebrae, articulation of these with the skull/first vertebra: nonflexible (0); flexible, vertebrae linked laterally and ventrally by ligamentous bands, which appear as ventral straps by transmitted light in lateral view (1).
42. First vertebra, dorsal extension of the neural spine when the neural arch and spine are plastered to the skull: the neural spine extending only slightly, or not at all, dorsally above its attachment to the skull (0); the neural spine with a long dorsal portion free from the skull beyond the curvature of the supraoccipital and the exoccipitals (1); not applicable (‘–').
43. Baudelot's ligament, placement of the proximal attachment: to the basioccipital (0); to the first vertebra (1); to the exoccipitals (2).
44. Neural spines, orientation: the neural spines of all (or all but the first few) of the abdominal vertebrae are orientated posterodorsally (0); several of the neural spines of the posterior abdominal and/or anterior caudal vertebrae are orientated anterodorsally, or at least vertically (1).
45. Haemal arch and spine, vacuities: no prominent vacuities (0); vacuities of moderate size present in the haemal arches or spines (primarily in the arches) of many of the posterior abdominal vertebrae and often present in those of the more anterior caudal vertebrae (1); vacuities of large size present in the haemal arches or spines of many of the posterior abdominal vertebrae and often present in those of the more anterior caudal vertebrae (2).
46. Abdominal haemal spines: many of the haemal spines of the abdominal vertebrae, especially posteriorly, with a prominent process in the midline below the bridge under the haemal canal (0); the haemal arches with a transverse bony bridge below the haemal canal, but without a median spine below the bridge, although short vertical projections may occur below the bridge on each side (1).
47. Ossified ribs: present on most of the abdominal vertebrae behind the fourth (0); present only on the last few abdominal vertebrae (1); present only on a few of the middle abdominal vertebrae (2); absent (3); present on all of the abdominal vertebrae except the first (4); present on all of the abdominal vertebrae except the first two (5).
48. Ossified epineurals: present on most of the abdominal vertebrae or their ribs (0); present only on a few of the anterior abdominal vertebrae (1); present only on a few of the middle abdominal vertebrae (2); no ossified epineurals (3).
49. (20) PU2, length of the neural spine: long (0); absent to short (1).
50. (21) Hypurapophysis: present (0); absent (1).
51. (22) Epurals, number: three (0); two (1); one (2).
52. Parhypural, articulation of the proximal end to the urostylar centrum: strongly embraces the centrum (0); slightly removed from and not embracing the centrum (1); laterally expanded as a specialized peg, with the pegs on each side of the parhypural fitting into sockets on each side of the centrum (2).
53. (24, 25) Hypurals, degree of fusion (+ indicates fused together): 4–6 separate hypural elements (0); hypurals 1 + 2 are fused together and to the centrum, and hypurals 3 + 4 are fused together and free from the centrum (1); hypurals 1 + 2 and hypurals 3 + 4 are fused to one another and to the centrum (2); hypurals 1 + 2 and hypurals 3 + 4 are separate from one another, and both plates are free from the centrum (3); hypurals 1 + 2 are free from the centrum, and hypurals 3 + 4 + 5 are either free or fused to the centrum (4); all the hypurals are fused to the centrum, and hypural 5 is not free (5).
54. (26) Uroneural: present (0); absent (1).
55. Stegural process (sensu Rosen, 1984; Grande et al., 2013): present (0); absent (1).
56. (23) PU2, extra-caudal ossicle between HPU2 and HPU3 (sensu Fujita, 1990; Grande et al., 2013): absent (0); present, in at least some specimens (1).
57. Dorsal-fin spine, locking mechanism, base of one spine against another: absent (0); present between the first and second dorsal-fin spines (1); present between the second and third dorsal-fin spines (2); present between the first, second, and third dorsal-fin spines (3); not applicable, when no dorsal-fin spines (‘–').
58. Vacant interneural spaces, number of groups (when two or more spaces are vacant): one (0); two (1); three (2); four (3); not applicable, when only one space or none vacant, or no spiny dorsal fin (‘–').
59. Dorsal-fin pterygiophores, number anterior to the neural spine of the fourth abdominal vertebra: none (0); two (1); three (2); four (3); five (4); not applicable, when no spiny dorsal fin (‘–').
60. First pterygiophore of the spiny dorsal fin, placement: behind the first interneural space, i.e., behind the second or subsequent neural spines (0); inserted in the first interneural space, i.e., between the first and second neural spines, or into what would be the preneural space if the first neural arch and spine were not plastered onto the skull, and often slanted forward (1).
61. (16) First dorsal-fin pterygiophore, position of the base in the first interneural/preneural space: middle to rear, not in contact with the skull and the neural arch and spine of the first vertebra (0); front, or fills the space, in contact with the skull and first vertebra between the two sides of the neural arch and spine of the first vertebra (1).
62. Spinous dorsal fin, distal radials: large, ossified (0); reduced, absent, or cartilaginous (1); not applicable, when no spiny dorsal fin present (‘–').
63. Soft dorsal- and anal-fin pterygiophores: asymmetrical (0); symmetrical (1).
64. (17) Supraneurals, number: none (0); one (1); two (2); three (3).
65. Supraneural, cartilage at the distal end: present (0); absent (1); not applicable, when no supraneurals present (‘–').
66. Anal-fin spine, locking mechanism, base of one spine against another: absent, when two or more spines are present (0); present between the first and second spines (1); not applicable, when a single or no anal spines present (‘–').
67. First anal-fin spine, articulation with the pterygiophore: unfused (0); fused in some populations or at larger specimen sizes (1); fused in all specimens (2); not applicable, when anal spines absent (‘–').
68. Second anal-fin spine, length: moderate to long, more than one-half the length of the first spine (0); short, less than one-half the length of the first spine (1); not applicable, when second anal spine absent (‘–').
69. Anal-fin pterygiophores, number of in the prehaemal space (anterior to the haemal spine of the first caudal vertebra): three (0); two (1); one (2); none (3).
70. Anal-fin pterygiophores, number in the first interhaemal space (between the haemal spines of the first and second caudal vertebrae): none (0); one (1); two (2); three (3).
71. Anal-fin pterygiophores, number in the second interhaemal space (between the haemal spines of the second and third caudal vertebrae): one (0); two (1); three (2).
72. Anal-fin pterygiophores, number anterior to the haemal spine of the third caudal vertebra: three (0); four (1); five (2); six (3); seven (4); eight (5); nine or more (6).
73. (15, 19) Dorsal-, anal-, and pectoral-fin rays: branched (0); unbranched (1).
74. Pectoral-fin radial, lateral flange on the lowermost: absent (0); present (1).
75. Postcleithrum, number of separate bony elements: two (0); one (1).
76. Single postcleithrum, flange: flange absent on the single postcleithrum (0); a flange present along the posterior border of the single postcleithrum, and the flange may be laterally flared (1); not applicable, when two postcleithra present (‘–').
77. Supracleithral serrations: none (0); serrations present along the posterior border, and this border sometimes laterally flared (1).
78. Supracleithrum, ventral end: simple (0); deeply bifurcate (1).
79. Cleithrum, posterior edge: without a posterodorsal prong above the articulation with the postcleithrum (0); cleithral process present as a prong above the articulation with the postcleithrum (1).
80. Extrascapulars: one long bone, sometimes forming an open tube, more or less closely held to the skull and integrated in line with the crest (often spiny) between the post-temporal and the parietal (0); two tubular bones, not closely held to the skull, except at larger specimen sizes (1); three more or less tubular bones (2).
81. Pelvic fin, position: approximately midway between the anus and the pectoral-fin base (0); slightly behind the pectoral-fin base (1); under or anterior to the pectoral-fin base (2); not applicable (‘–').
82. Pelvic-fin spines: present (0); absent (1); not applicable (‘–').
83. Pelvic-fin rays, anterolateral processes of the medial (lower) surfaces: absent (0); present as prongs from the medial surfaces of the ray bases (1); present as broad flanges from the ray bases (2); not applicable (‘–').
84. Pelvic-fin rays, serrations: absent (0); present on crests on the anterior or upper and/or the lower posterior surfaces of several rays (1); present on broad flanges from the medial (lower) surface of several rays (2); not applicable (‘–').
85. Basipterygia, articulation: the medial processes of the basipterygia broadly overlap at the level of the pelvic fin (0); in contact in the midline of the middle region, but with little or no overlap (1); not in close contact in the middle region, although often in contact at the anterior ends (2); tightly adherent or partially fused along a broad area of midline contact (3).
86. Pelvis, posterior process behind pelvic-fin base: short to moderate in length, and in shape a moderate to broad plate or flattened shaft, usually slightly to distinctly obliquely orientated, with or without flanges and retrorse projections (0); long and rod-like, moderately separated from its opposite member along the midline (1); not applicable (‘–').
87. Scales, on most of the body: moderate to small, spiny ‘ctenoid' (spinoid) scales (0); moderate to small, cycloid scales (1); scales greatly elongate vertically (2); scales absent (excluding enlarged buckler-like scales), or with only lateral line scales (3); not applicable (‘–').
88. Scales, buckler-like (greatly enlarged midline scales): absent (0); present only from the isthmus to the anus (1); present midabdominally and from the rear end of the spinous dorsal fin to the end of the soft dorsal-fin base (2); present midabdominally and from below the spinous dorsal-fin base (usually from the front to middle region) to the end of the soft dorsal-fin base (3).
89. Scales, scute-like (slightly enlarged midline scales): absent (0); present from the isthmus to the pelvic-fin base, and sometimes more posteriorly (1).
90. Scales, along the bases of the dorsal- and anal-fin rays: present along the bases of the fin rays, usually as a low sheath of scales that lack spiny processes (0); absent from the bases of the rays, and the scales nearby without spiny projections and not extending beyond the lateral expansions of the distal ends of the dorsal- and anal-fin pterygiophores (1); absent along the bases of the rays, but spiny processes present on the scales alongside the lateral expansions of the distal ends of the dorsal- and anal-fin pterygiophores (2).
91. Hyperostosis: absent (0); present in the supraoccipital and the first dorsal-fin pterygiophore of some specimens (1); present in the prepelvic scale bucklers (2).
92. (1) “Gadoid notch”: absent (0); present (1).
93. Vertebrae, total number: 26 or fewer (0); 27 or 28 (1); 29 or 30 (2); 31 or 32 (3); 33 or 34 (4); 35 or 36 (5); 37 or 38 (6); 39 or 40 (7); 41 or 42 (8); 43 or more (9).
94. Abdominal vertebrae, number: nine or fewer (0); 10 (1); 11 (2); 12 (3); 13 (4); 14 (5); 15 (6); more than 15 (7).
95. Vertebrae, number in the caudal peduncle (posterior to the last vertebra whose neural or haemal spine supports a pterygiophore): three (0); four (1); five (2); six (3); seven (4); eight (5); nine (6); 10 (7); 11 or more (8).
96. Principal caudal-fin rays, number: 16 or more (0); 15 (1); 14 (2); 13 (3); 12 (4); 11 (5).
97. Procurrent caudal-fin rays, number (including the number in both the dorsal and ventral sides, if different): none (0); one (1); two (2); three (3); four (4); five (5); six (6); seven (7); eight (8); nine or more (9).
98. Dorsal-fin spines, number: four or fewer (0); five (1); six (2); seven (3); eight (4); nine (5); 10 or more (6); not applicable, when no spiny dorsal fin present (‘–').
99. Vacant interneural spaces, total number below the spiny and anterior part of the soft dorsal-fin base, posterior to the first dorsal-fin pterygiophore: none (0); one (1); two (2); three (3); four (4); five (5); seven (6); eight (7); nine (8); not applicable, when no spiny dorsal fin present (‘–').
100. Anal-fin spines, number: none (0); one (1); two (2); three (3); four (4).
101. Pectoral-fin rays, number: 0–16 (0); 17 and above (1).
102. Pelvic-fin elements, total number: nine (0); eight (1); seven (2); six (3); five (4); four (5); three (6); one (7).
103. (12) Branchiostegal rays, 7 or more (0); 6 or fewer (1).
104. (2) Supramaxillae: both present (0); both absent (1).
105. (4) Shape of opercle: oval or rounded (0); diamond shaped (1).
106. (5) Spatial relationship of levator arcus palatine to section A2 the adductor mandibulae: medial or does not connect to the abductor mandibulae complex (0); lateral to the dorsal border of section A2 of the abductor mandibulae complex (1).
107. (6) Number of hyomandibular condyles: 2 (0); 1 (1).
108. (7) Size of intercalar: small (0); enlarged (1).
109. (8) Exoccipital facets: close together (0); widely separated (1).
110. (9) Otolith shape: oblong (0); pince-nez shaped (1).
111. (10) Basihyal: present (0); lost (1).
112. (13) Percopsoid projections: absent (0); present (1).
113. (18) Scapular foramen: bounded by scapula (0); not bounded solely by scapula (1).
114. Lower process of hyomandibula (Endo, 2002): absent (0); present (1).
115. (Borden 22) Flexor ventralis externus: absent (0); present (1).
116. (Borden 23) Flexor ventralis serves at least on principal ray in dorsal series: no (0); yes (1).
117. (Borden 24) Majority of ural centra and preural centra 2–4 exposed after removal of body musculature: no (0); yes (1).
118. (Borden 15) Location of interradialis: across and between rays (0); between fin rays only (1).
119. (Borden 16) Insertion site of interradialis: principal caudal rays only (0); principal caudal rays plus procurrent and/or anal-fin/dorsal-fin rays (1).





