GigaDB - Submission Guidelines
General Submission Guidelines
GigaDB is a China National GeneBank supported repository used to host data and tools associated with articles in GigaScience. As part of your manuscript submission and in line with the Reporting Standards and FAIRsharing guidelines for data deposition and formatting for papers submitted to GigaScience we will provide an associated GigaDB dataset to host the data and files required for transparency and reproducibility. GigaDB is an open-access database. As such, all data submitted to GigaDB must be fully consented for public release (for more information about our data policies, please see our Terms of use page).
Workflow
The workflow diagram below details a standard submission process:
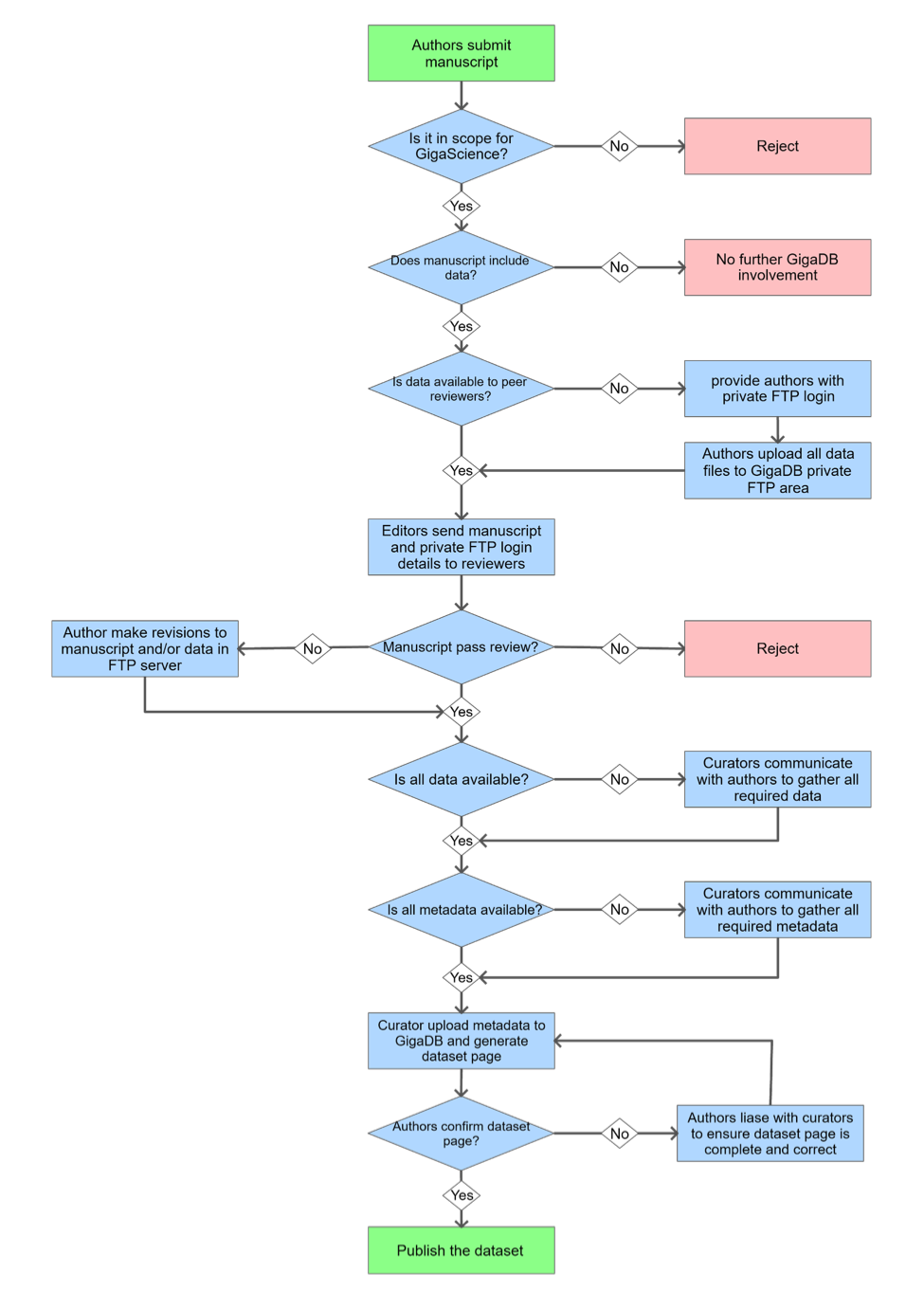
Workflow overview
This workflow diagram outlines the manuscript and data submission process for GigaScience. It covers the steps from initial manuscript submission to the eventual publication of the dataset.
Workflow Steps
- Authors submit manuscript
- Is it in scope for GigaScience?
- Decision: If no, reject. If yes, continue.
- Does manuscript include data?
- Decision: If no, no further GigaDB involvement. If yes, continue.
- Is data available to peer reviewers?
- Decision: If no, provide authors with private FTP login, then authors upload all data files to GigaDB private FTP area and continue. If yes, continue.
- Editors send manuscript and private FTP login details to reviewers
- Does manuscript pass review?
- Decision: If no, either reject or author makes revisions to manuscript and/or data in FTP server, and continue. If yes, continue.
- Is all data available?
- Decision: If no, gather all required data. If yes, continue.
- Is all metadata available?
- Decision: If no, gather all required metadata. If yes, continue.
- Curator uploads metadata to GigaDB
- Did authors confirm dataset page?
- Decision: If no, authors liaise with curators to ensure dataset page is complete and correct, then again curators upload metadata to GigaDB and generate dataset page. If yes, publish dataset.
When contacted by curators to process the GigaDB dataset you will be invited to:
- Create a GigaDB user account
- Upload your prepared data files if not already public (see checklists below)
- Supply the appropriate metadata
- Proofread and approve the GigaDB pre-publication dataset page
Required metadata
For all datasets the following information will be required. Most of the details will be imported directly from the GigaScience manuscript submission, other details will be requested by the curators.
For datasets that include biological sample-related data we would expect the sample metadata to be included in the GigaDB dataset. We understand that the level of sample metadata made available is often limited by sample collection restrictions, but authors should make every effort to provide as comprehensive metadata about samples as is possible.
Below is the list of attributes commonly associated with any biological sample. In addition to these we strongly encourage the inclusion of ALL appropriate attributes, and for specific types of data there are a number of standards that we encourage our users to adopt. Please see the Dataset Type specific checklists for recommendations.
For all datasets we expect all data to be available from a stable public open access source and where appropriate we will link directly to external sources rather than duplicate data files.
However if there is no established suitable repository for a particular file/data-type we will host it on our servers.
Where possible, all files should be machine readable without the need for proprietary software (e.g. No PDF, Excel or Word documents).
For all files we host, we expect the following details:
Due to the nature of scientific publications the files that need to be provided are usually unique to the individual manuscript, however there are some commonalities that we have attempted to capture in a set of minimal checklists for the most common dataset types that we receive. These lists are to be treated as a guide only and there may be changes to them over time.
Please see the Dataset Type specific checklists for recommendations:
If you have any questions, please contact us at database@gigasciencejournal.com.
