Dear Colleagues,
MicroRNAs (miRNAs) are single-stranded, small non-coding RNA molecules that epigenetically regulate gene expression, mostly through binding to the 3′ untranslated region (3′ UTR) of targeted messenger RNA (mRNA) molecules and, hence, mediating translational repression, usually combined with or followed by mRNA degradation. The human genome can produce more than a thousand of different miRNAs, which regulate approximately 70% of protein-coding genes, thereby controlling the activity and function of key signaling pathways and cellular processes such as cell proliferation, apoptosis, cell differentiation, and response to hypoxia. In fact, miRNAs can function either as oncogenes or tumor suppressors. From a clinical perspective, particular miRNAs can also serve as molecular biomarkers and therapeutic targets for several human diseases.
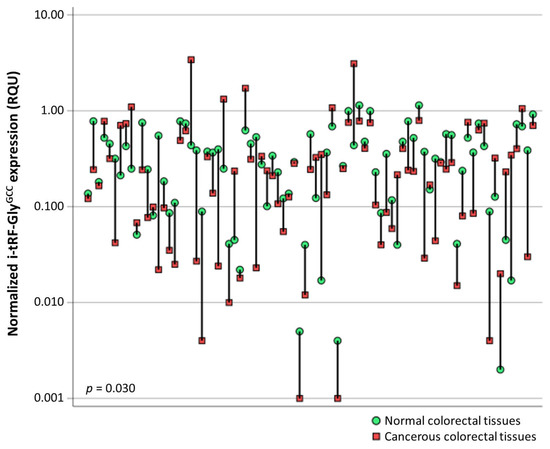
tRNA-derived RNA fragments are small non-coding RNAs specifically cleaved from transfer RNA (tRNAs), with a length of 14–48 nucleotides. They are classified into three main classes, namely the tRNA fragments (tRFs), the stress induced tRNA derived RNA fragments (tiRNAs), and the toxic small tRNA derived RNA fragments (tsRNAs). The tRNA molecules present a high differentiation in their cleavage sites, thus leading to several tRNA-derived RNA fragments varying in length and having distinct functions. They participate in translation regulation and gene silencing with a subsequent effect on cell viability and proliferation. Furthermore, they play a pivotal role in stress-induced situations.
Circular RNAs (circRNAs) are a neglected RNA type deriving from back-splicing. Initially, they were characterized as by-products of alternative splicing. However, the high-throughput analysis revolution uncovered their widespread expression, arousing the scientific interest. circRNAs have been designated as crucial modulators in several aspects of cell life, both in physiological and pathological states. CircRNAs are heavily involved in biological processes by acting either as sponges of miRNAs and RNA-binding proteins (RBPs) or by encoding for peptides. Especially, their miRNA-sponging activity implies that circRNAs can affect post-transcriptional gene regulation mediated by miRNAs. As studies regarding the intricate functions of circRNAs emerged, their association with multiple regulatory networks became evident, proving that these molecules play key roles in human disease development and may have wide clinical application.
Long noncoding RNAs (lncRNAs) are abundantly transcribed by the human genome. During the last decade, it has become evident that mutations and dysregulation of expression of lncRNAs significantly contribute to many human diseases. Alterations in the primary structure, secondary structure, and expression levels of lncRNAs as well as their cognate RNA-binding proteins have been shown to underlie the pathobiology of cancer, neurodegenerative, and cardiovascular diseases. Thus, recent scientific progress suggests that the involvement of lncRNAs in human diseases could be far more prevalent than previously thought.
This topic collection focuses on the information of the readers regarding the potential of miRNAs, tRNA-derived RNA fragments, circRNAs and other long non-coding RNAs to regulate protein-coding gene expression at a post-transcriptional level, in human physiological and pathological states. The authors are encouraged to submit their original research studies concerning this topic. Review articles will also be taken into consideration. We hope that this topic collection regarding the identity, biological role, and/or clinical utility of miRNAs, tRNA fragments, lncRNAs and circRNAs will arise the interest of the readers of this journal.
Dr. Christos K. Kontos
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomedicines is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.