Open
Description
Describe the workflow you want to enable
I wanted to ask whether any plans exist to implement a rolling/sliding window method in the TimeSeriesSplit class:
Currently, we are limited to using the expanding window type. For many financial time series models where a feature experiences a structural break, having a model whose weights are trained on the entire history can prove suboptimal.
I noted in #13204, specifically svenstehle's comments, that this might be on the horizon?
Describe your proposed solution
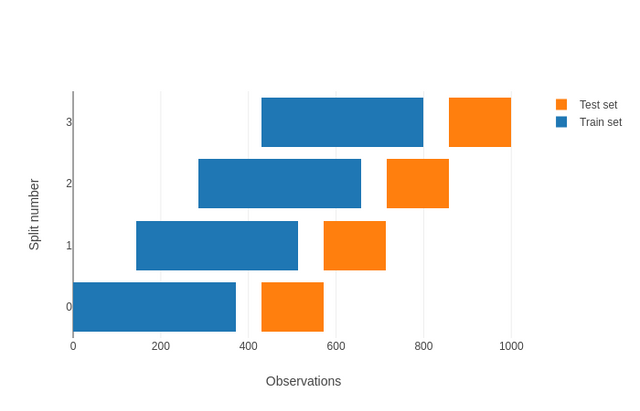
Current Implementation
>>> x = np.arange(15)
>>> cv = TimeSeriesSplit(n_splits=3, gap=2)
>>> for train_index, test_index in cv.split(x):
... print("TRAIN:", train_index, "TEST:", test_index)
TRAIN: [0 1 2 3] TEST: [6 7 8]
TRAIN: [0 1 2 3 4 5 6] TEST: [ 9 10 11]
TRAIN: [0 1 2 3 4 5 6 7 8 9] TEST: [12 13 14]
Desired outcome
>>> x = np.arange(10)
>>> cv = TimeSeriesSplit(n_splits='walk_fw', max_train_size=3, max_test_size=1)
>>> for train_index, test_index in cv.split(x):
... print("TRAIN:", train_index, "TEST:", test_index)
TRAIN: [0 1 2] TEST: [3]
TRAIN: [1 2 3] TEST: [ 4]
TRAIN: [2 3 4] TEST: [5]
TRAIN: [ 3 4 5] TEST: [6]
TRAIN: [ 4 5 6] TEST: [7]
TRAIN: [ 5 6 7] TEST: [8]
TRAIN: [ 6 7 8] TEST: [9]
Where the 'stride' of the walk forward is proportionate to the test set, or walks by the max_train_size parameter?
Describe alternatives you've considered, if relevant
No response
Additional context
No response