-
-
Notifications
You must be signed in to change notification settings - Fork 26.5k
Description
Dear Scikit team,
I really enjoy using the scikit learn library and I am very greatful for your very impactful work. I hope I can contribute with this bug report (if it is true) to this library.
Keep on your good work!
I checked existing issues that concern MDS it is not the same as #16846 but to solve it I would also address #11381
Describe the bug
Everything concerns the MDS implementation in scikit-learn/sklearn/manifold/_mds.py. I have copied the relevant snipped of code below and added the line numbers I am referring to. All equations I am referring to are from [1]
Short version:
- The initialization of the disparities in line 103 leads to an error when calculating the stress and the update of the configuration.
- The standardization in line 106 is wrong.
- The library can not reproduce results from the r library mass
Long version:
I found the following error in the implementation:
-
In
scikit-learn.sklearn.manifold._mds._smacof_singlein line 103 the disparities get initialized with the distances. In line 104 only the values of the upper triangle matrix get overwritten by the calculated disparities (disparities_flat). This results in a matrix that has disparities (similar to rank distances) in the upper triangle and euclidean distances in the lower triangle. Therefore the normalization factor in line 106 is calculated based on a matrix that is half distances and half disparities -
Line 106 standardized the disparities to
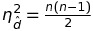
which corresponds to step 3 and 9 of theSmacof algorithm with Admissibly Transformed Proximities[2]. However, from equation 9.1 it seems like this corresponds only to the upper triangle matrix (which explains then(n-1)/2) in the implementation of line 106, all elements from the matrix are used which results in a wrong standardization factor. -
Also line 106 the term
(disparities ** 2).sum()should not be in the square-root. This results from a transformation when we are looking for a normalization factorathat is applied to the disparities such that:
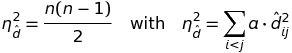
We then should calculateaas
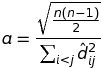
(equations 9.1 + 9.2) from [1] -
When the stress is computed in line 110, a difference between the dissimilarities and the disparities is computed. As stated above, the disparities had the dissimilarities in the lower triangle matrix (which I think is confusing) which would normally mean that the all differences in the lower triangle matrix are zero. However, as the disparities got scaled in line 106, the differences in the lower triangle matrix are no longer zero but some (more or less) random values.
There are also some smaller issues that I noticed:
- After the last update of the configuration, the stress is not updated. This means the old stress is returned (I think this is the same as In multifold.MDS stress value doesn’t correspond to returned coordinates #16846)
- I think it would be good to return the normalized stress
References
[1]: “Modern Multidimensional Scaling - Theory and Applications” Borg, I.; Groenen P. Springer Series in Statistics (1997), Version from 2005
[2]: “Modern Multidimensional Scaling - Theory and Applications” Borg, I.; Groenen P. Springer Series in Statistics (1997), Version from 2005, Page 204 in Chapter 9. Metric and non-metric MDS
Code from scikit MDS implementation (for reference)
ir = IsotonicRegression()
for it in range(max_iter):
# Compute distance and monotonic regression
dis = euclidean_distances(X)
if metric:
disparities = dissimilarities
else:
dis_flat = dis.ravel()
# dissimilarities with 0 are considered as missing values
dis_flat_w = dis_flat[sim_flat != 0]
# Compute the disparities using a monotonic regression
disparities_flat = ir.fit_transform(sim_flat_w, dis_flat_w)
[103] disparities = dis_flat.copy()
[104] disparities[sim_flat != 0] = disparities_flat
[105] disparities = disparities.reshape((n_samples, n_samples))
[106] disparities *= np.sqrt((n_samples * (n_samples - 1) / 2) /
(disparities ** 2).sum())
# Compute stress
[110] stress = ((dis.ravel() - disparities.ravel()) ** 2).sum() / 2Steps/Code to Reproduce
The code and data to calculate the results is in this gist: (https://gist.github.com/henrymartin1/05a2aa5210e52216abd7ce61e6918e3a). The relevant file isscikit_mds_bugreport.py
I honestly had a hard time to come up with code that proofs that there is a problem. This is what I did:
- I created an array and its distance matrix in python and stored it as .csv
- Using this data, I calculated the optimal configuration and the stress using the non-metric MDS implementation of the r-library mass
- I calculate the optimal configuration and the stress using scikit-learns mds
- I calculate the non-metric stress using my own implementation of non-metric stress
- I plot the optimal scikit configuration and the optimal r configuration
Expected Results
- The two optimal configurations would be the same or very similar
- The stress reported by r and the stress reported by scikit are the same
- The configurations found by scikit and by r have the same stress when I calculate it using my own implementation of the stress function. (+ they match with what is reported by scikit and the r implementation)
Actual Results
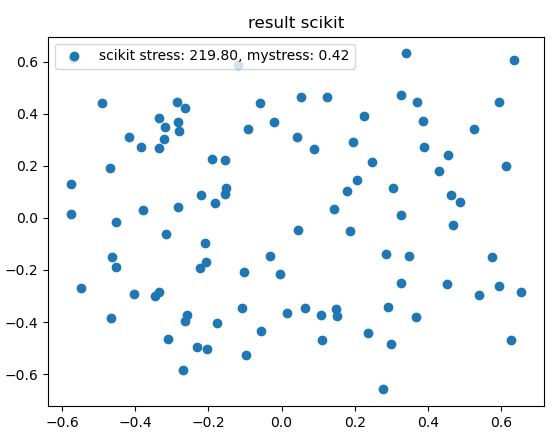
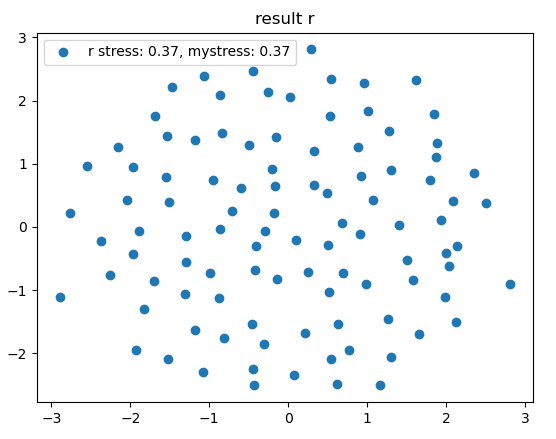
- The optimal scikit configuration and the optimal r configuration are significantly different (see plots below)
- My calculated stress for the r configuration and the scikit configuration are different
- The scikit stress is not normalized (not necessarily a problem)
Result mds scikit
Result mds r
Versions
System:
python: 3.8.6 | packaged by conda-forge | (default, Oct 7 2020, 18:22:52) [MSC v.1916 64 bit (AMD64)]
executable: C:\Users\henry.conda\envs\evhomepv\python.exe
machine: Windows-10-10.0.19041-SP0
Python dependencies:
pip: 20.2.4
setuptools: 49.6.0.post20201009
sklearn: 0.23.2
numpy: 1.19.4
scipy: 1.5.3
Cython: None
pandas: 1.1.4
matplotlib: 3.3.2
joblib: 0.17.0
threadpoolctl: 2.1.0
Built with OpenMP: True