-
-
Notifications
You must be signed in to change notification settings - Fork 26.7k
Open
Labels
Description
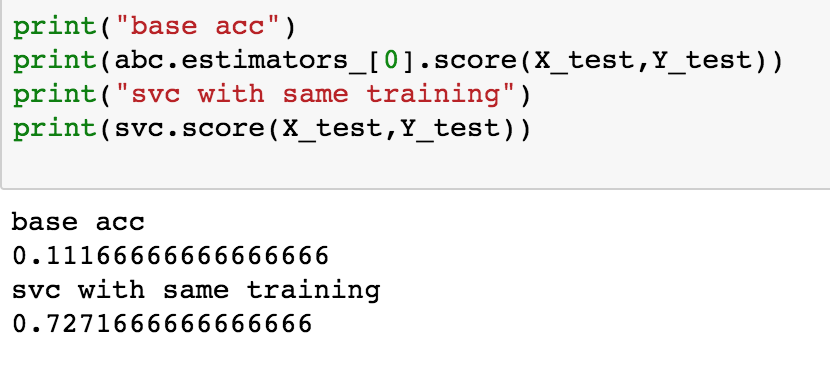
I am trying to use SVC with ADAboosting. WIth SVC, but not other base estimators, the initial estimator does not seemed to be trained.
Comparing the initial estimator of the ensemble to a single estimator with the same hyper-parameters
create an ADA ensemble
Create a single SVC
Compare the accuracies
I have tried different hyper-parameters and had the same issue. This issue does happen for RandomForests and DecisionTree Classifiers
Reactions are currently unavailable