-
Notifications
You must be signed in to change notification settings - Fork 594
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
merge plot_inverse_covariance_connectome and plot_multi_subject_connectome #1223
Comments
|
I'd rather not rename plot_simulated_connectome, as I don't see the need to commit to sparse estimators for the example on simulated connectomes. With regards to the other examples, agreed that it's a mess. Indeed the didactic values of the 2 examples are not well separated. Also, there is plot_signal_extraction that goes a bit in the same direction, even though it is on a hard parcellation, rather than a probabilistic atlas. They bring a variety of teaching:
I wonder if it's a good thing to put in the same example the single subject and the group analysis. I would rather merge the one with the probabilistic parcellation and the one with the hard parcellation. We should rename the multi-subject one to a name that makes it very clear that it's purpose is to illustrate multi-subject. For instance a file name "plot_multi_subject_connectomes" and a title "Computing connectomes on multiple subjects". |
|
Ok. |
|
What I can do for all examples (except ICA and Dictionary learning, cause
I am a stranger to that) is to synthesize in a table what they demo piece
by piece, to see usefulness and overlaps.
I think that this would be very useful. First for our internal
refactoring, but after to help explain better which example should be
read.
I also have a question: is there any reason we rely only on ADHD for
examples ? for advanced connectomes examples we can rely on COBRE/ABIDE
to avoid the signal extraction part
Well, this part is useful to teach to people. I like the idea that we
always start from 4D fMRI files, as this is what people have.
In order to limit the downloads, I'd like all the examples to rely on the
same 4D fMRI files, as these are very heavy. However, in the long run, I
think that I would like to try to have all examples running with the
cobre data. One of the reasons is that the prediction example would then
work much better. But I don't know how realistic it would be: would all
the examples work well, or not? Also, it seems to me that subjects take a
bit more space with Cobre than with ADHD: 65Mb on average per subject for
Cobre, and 45Mb for ADHD.
|
You would keep ADHD only to demo signal extraction ? |
|
You would keep ADHD only to demo signal extraction ?
No: I'd remove fully ADHD. But once again, I am a bit worried that this
might use too much disk space for the continuous integration server.
|
|
AFAIK, ADHD dataset is more used than COBRE and there has been a competition on it so people in the field are more likely to know it. |
|
AFAIK, ADHD dataset is more used than COBRE and there has been a competition on
it so people in the field are more likely to know it.
Yes, but it has nothing in it: it's very hard (impossible?) to predict
anything from it.
|
|
I did a table with the connectivity examples, except those ICA and dictionary learning related. I added a secondary teaching box to group demoed features/explanations that may be additional to the example purpose. I tried to separate nilearn API specific teaching from more general functional connectivity teaching, because some users can be beginners in FC. |
|
So from what I read I have some little changes suggestions
2- In section 3.2.Connectome extraction: inverse covariance for direct connections adding once for all an explanation for the link covariance/correlation/precision/partial correlation. 3- Standardization is a hidden process to use covariance as a correlation. May be we can expose 4- following previous discussions, decide whether or not 5- fix ADHD changing TR across the examples 6- Why systematically use the first figure in gallery ? For instance for the seed-to-voxel correlations it would be beautiful to see the correlation map, instead of a timeseries. 7- We can merge probabilistic/determinsitic/DMN spheres examples, it would be an example with all the various maskers. and I have a question: what are the guidelines for examples writing style ? For the moment they are a mix of compact minimalistic phrasing (eg the probabilistic atlas extraction) and more detailed structured examples (eg the seed-to-voxel example). |
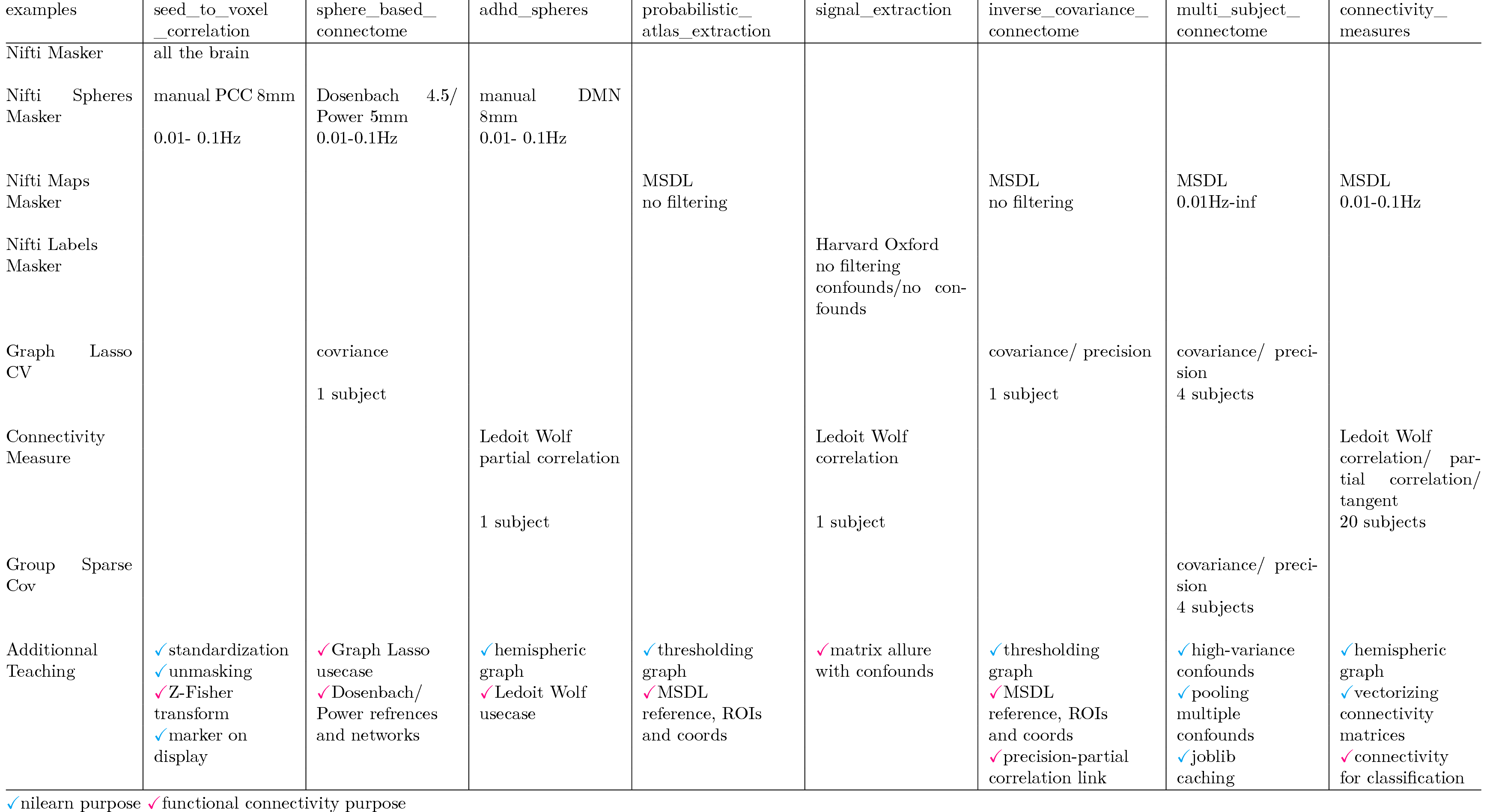
The connectivity example
plot_inverse_covariance_connectomedemossklearn.covariance.GraphLassoCVon ADHD data with MSDL atlas. The exampleplot_multi_subject_connectomecompares the GraphLasso tonilearn.connectome.GroupSparseCovarianceCValso for ADHD/MSDL.Morevover, we have the same comparison on simulated data in
plot_simulated_connectomeexample. I was thinking of:plot_inverse_covariance_connectomeplot_multi_subject_connectome->plot_sparse_connectomesplot_simulated_connectome->plot_simulated_sparse_connectomes3. adding an example
plot_connectome_estimatorsto demo the different covariance estimators: EmpiricalCovariance, LedoitWolf and GraphLassoThe text was updated successfully, but these errors were encountered: